Rearranged Oriloc output example: Previous Index Next
Bacteria; Firmicutes; Lactobacillales; Enterococcaceae; Enterococcus.
Genome size (bp): 3218031
Number of genes 3113
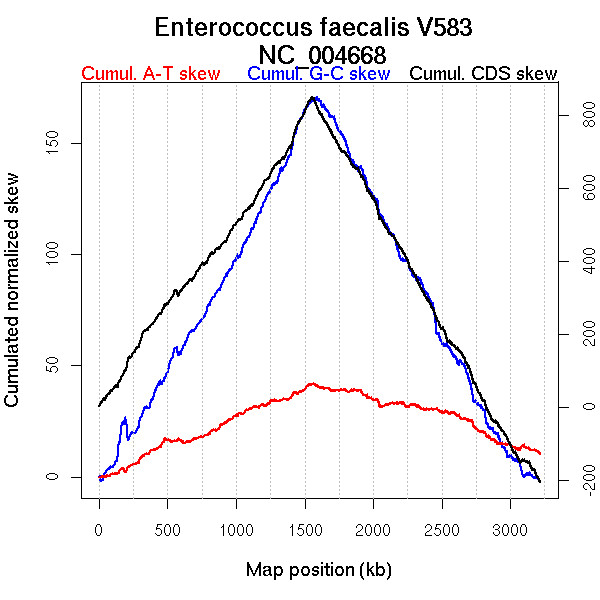
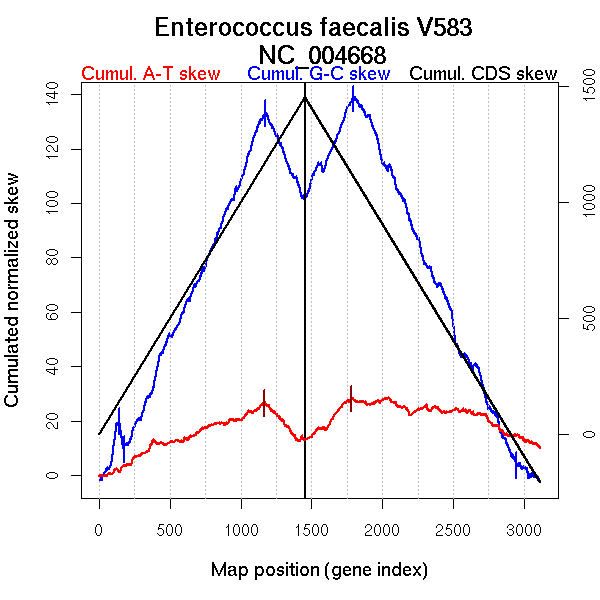
Oriloc predictions: Origin 0 kb Terminus 1587 kb
Worning et al., 2006: Origin 3217 kb Terminus 1551 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 1.449 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 0.73 kb
Consensus predictions: Origin 0 kb Terminus 1587 kb
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 140 | 0 | 190 |
| 175 | 0 | 213 | |
| 1172 | 0 | 1549 | |
| GC-skew reverse | 1792 | 0.00667 | 1577 |
| 2946 | 0.04667 | 2979 | |
| AT-skew forward | 1167 | 0 | 1543 |
| AT-skew reverse | 1783 | 0 | 1563 |
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 140 (189.9815 kb) | leading | 0.164 |
| 141 (191.7025 kb) | 175 (212.876 kb) | leading | -0.316 | |
| 176 (214.2415 kb) | 1172 (1548.855 kb) | leading | 0.123 | |
| 1173 (1553.8395 kb) | 1453 (3217.567 kb) | lagging | -0.118 | |
| GC-skew reverse | 1454 (0 kb) | 1792 (1577.2375 kb) | leading | 0.104 |
| 1793 (1577.6035 kb) | 2946 (2979.2805 kb) | lagging | -0.121 | |
| 2947 (2980.752 kb) | 3113 (3217.567 kb) | lagging | -0.037 | |
| AT-skew forward | 1 (0 kb) | 1167 (1543.0155 kb) | leading | 0.022 |
| 1168 (1545.265 kb) | 1453 (3217.567 kb) | lagging | -0.054 | |
| AT-skew reverse | 1454 (0 kb) | 1783 (1562.545 kb) | leading | 0.05 |
| 1784 (1565.168 kb) | 3113 (3217.567 kb) | lagging | -0.012 |
More G than C on the leading strand for replication.