Rearranged Oriloc output example: Previous Index Next
Bacteria; Proteobacteria; Epsilonproteobacteria; Campylobacterales; Helicobacteraceae; Wolinella.
Genome size (bp): 2110355
Number of genes 2043
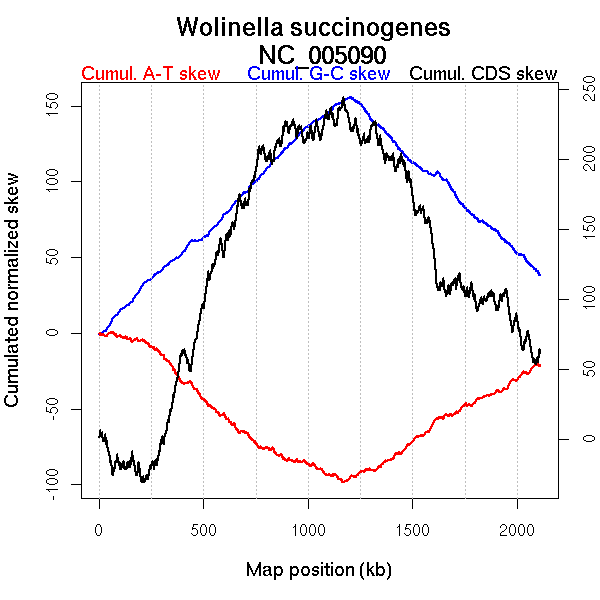
Oriloc predictions: Origin 2 kb Terminus 1188 kb
Worning et al., 2006: Origin 3 kb Terminus 1187 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 577.98 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 0.66 kb
Consensus predictions: Origin 2 kb Terminus 1188 kb
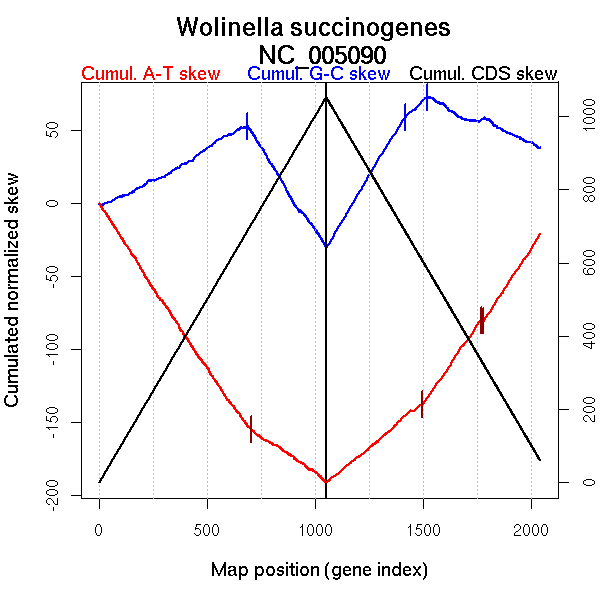
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 688 | 0 | 1186 |
| GC-skew reverse | 1420 | 0.04 | 987 |
| 1521 | 0 | 1206 | |
| AT-skew forward | 705 | 0 | 1241 |
| AT-skew reverse | 1498 | 0.04222 | 1173 |
| 1769 | 0 | 1610 | |
| 1781 | 0 | 1616 |
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 688 (1186.425 kb) | leading | 0.084 |
| 689 (1197.7575 kb) | 1052 (2110.227 kb) | lagging | -0.229 | |
| GC-skew reverse | 1053 (0 kb) | 1420 (986.7825 kb) | leading | 0.247 |
| 1421 (989.3985 kb) | 1521 (1206.2 kb) | leading | 0.137 | |
| 1522 (1209.098 kb) | 2043 (2110.227 kb) | lagging | -0.062 | |
| AT-skew forward | 1 (0 kb) | 705 (1241.0945 kb) | leading | -0.223 |
| 706 (1247.355 kb) | 1052 (2110.227 kb) | lagging | -0.099 | |
| AT-skew reverse | 1053 (0 kb) | 1498 (1173.144 kb) | leading | 0.124 |
| 1499 (1173.95 kb) | 1769 (1610.395 kb) | lagging | 0.215 | |
| 1770 (1611.039 kb) | 1781 (1616.155 kb) | lagging | -0.031 | |
| 1782 (1618.301 kb) | 2043 (2110.227 kb) | lagging | 0.225 |
More G than C on the leading strand for replication.