Rearranged Oriloc output example: Previous Index Next
Bacteria; Proteobacteria; Alphaproteobacteria; Rhizobiales; Bradyrhizobiaceae; Rhodopseudomonas.
Genome size (bp): 5459213
Number of genes 4812
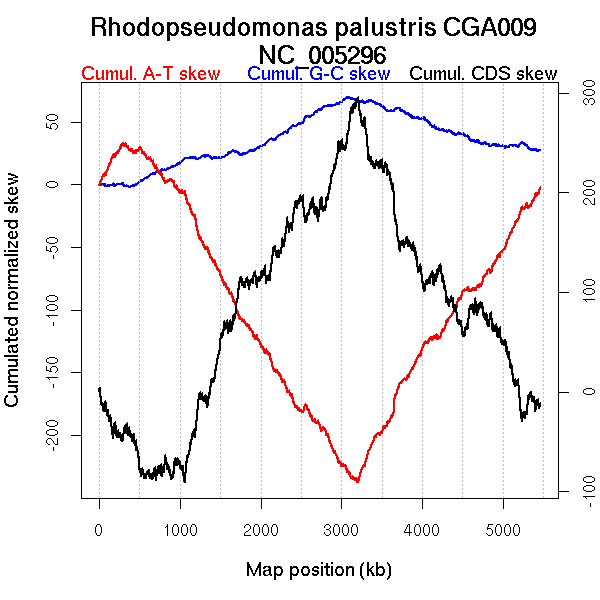
Oriloc predictions: Origin 366 kb Terminus 3093 kb
Worning et al., 2006: Origin 519 kb Terminus 3169 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 3968.681 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 1.39 kb
Consensus predictions: Origin 366 kb Terminus 3093 kb
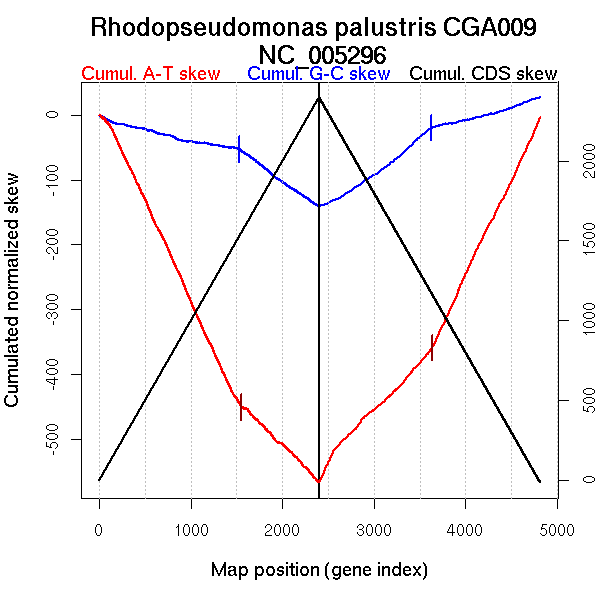
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 1526 | NA | 3151 |
| GC-skew reverse | 3033 | NA | 1403 |
| 3611 | NA | 3062 | |
| AT-skew forward | 127 | NA | 330 |
| 898 | NA | 1891 | |
| 1022 | NA | 2151 | |
| 1553 | NA | 3200 | |
| AT-skew reverse | 3631 | NA | 3136 |
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 1526 (3150.7445 kb) | leading | -0.032 |
| 1527 (3154.5955 kb) | 2399 (5458.655 kb) | lagging | -0.103 | |
| GC-skew reverse | 2400 (0 kb) | 3033 (1403.192 kb) | NA | 0.084 |
| 3034 (1406.8295 kb) | 3611 (3062.2235 kb) | leading | 0.123 | |
| 3612 (3067.299 kb) | 4812 (5458.655 kb) | lagging | 0.039 | |
| AT-skew forward | 1 (0 kb) | 127 (329.837 kb) | lagging | -0.15 |
| 128 (330.492 kb) | 898 (1890.5945 kb) | leading | -0.303 | |
| 899 (1891.754 kb) | 1022 (2150.715 kb) | leading | -0.342 | |
| 1023 (2152.338 kb) | 1553 (3200.3685 kb) | leading | -0.298 | |
| 1554 (3204.616 kb) | 2399 (5458.655 kb) | lagging | -0.138 | |
| AT-skew reverse | 2400 (0 kb) | 3631 (3135.7515 kb) | NA | 0.15 |
| 3632 (3136.391 kb) | 4812 (5458.655 kb) | lagging | 0.297 |
More G than C on the leading strand for replication - for forward encoded genes.