Rearranged Oriloc output example: Previous Index Next
Bacteria; Proteobacteria; Alphaproteobacteria; Rhizobiales; Bradyrhizobiaceae; Rhodopseudomonas.
Genome size (bp): 4892717
Number of genes 4397
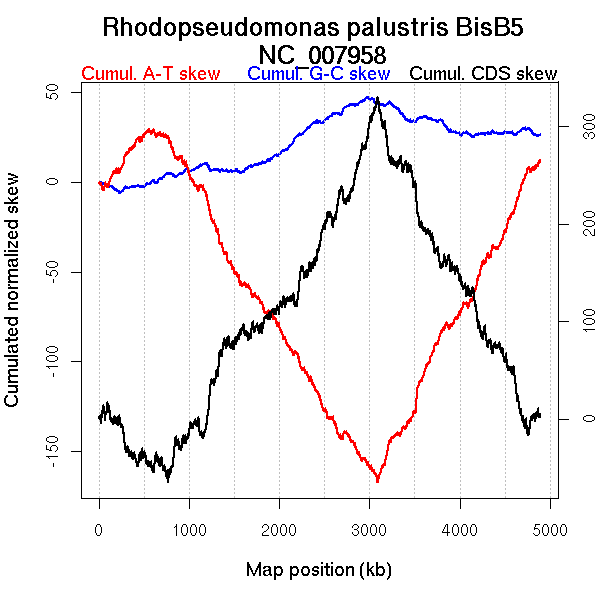
Oriloc predictions: Origin 234 kb Terminus 2963 kb
Worning et al., 2006: Origin 542 kb Terminus 3092 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 3747.063 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 1.36 kb
Consensus predictions: Origin 234 kb Terminus 2963 kb
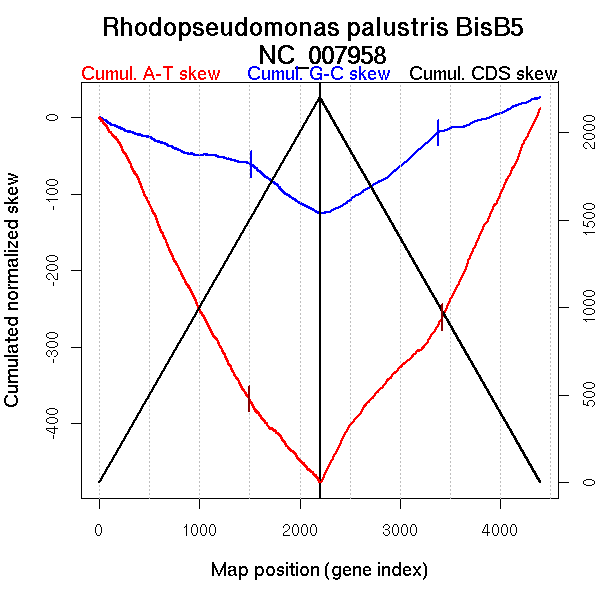
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 1514 | NA | 3047 |
| GC-skew reverse | 2867 | NA | 1596 |
| 3381 | NA | 2980 | |
| AT-skew forward | 281 | NA | 687 |
| 709 | NA | 1507 | |
| 1286 | NA | 2639 | |
| 1551 | NA | 3117 | |
| AT-skew reverse | 3421 | NA | 3098 |
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 1514 (3046.773 kb) | leading | -0.039 |
| 1515 (3048.847 kb) | 2200 (4892.034 kb) | lagging | -0.096 | |
| GC-skew reverse | 2201 (0 kb) | 2867 (1595.52 kb) | NA | 0.077 |
| 2868 (1601.8235 kb) | 3381 (2979.5295 kb) | leading | 0.116 | |
| 3382 (2982.9065 kb) | 4397 (4892.034 kb) | lagging | 0.047 | |
| AT-skew forward | 1 (0 kb) | 281 (687.235 kb) | NA | -0.182 |
| 282 (690.0505 kb) | 709 (1506.59 kb) | leading | -0.287 | |
| 710 (1511.552 kb) | 1286 (2639.112 kb) | leading | -0.249 | |
| 1287 (2639.823 kb) | 1551 (3117.4235 kb) | NA | -0.226 | |
| 1552 (3121.622 kb) | 2200 (4892.034 kb) | lagging | -0.144 | |
| AT-skew reverse | 2201 (0 kb) | 3421 (3098.4135 kb) | leading | 0.162 |
| 3422 (3100.591 kb) | 4397 (4892.034 kb) | lagging | 0.278 |
More G than C on the leading strand for replication - for forward encoded genes.