Rearranged Oriloc output example: Previous Index Next

Bacteria; Proteobacteria; Alphaproteobacteria; Rhizobiales; Bradyrhizobiaceae; Rhodopseudomonas.
Genome size (bp): 5331656
Number of genes 4683
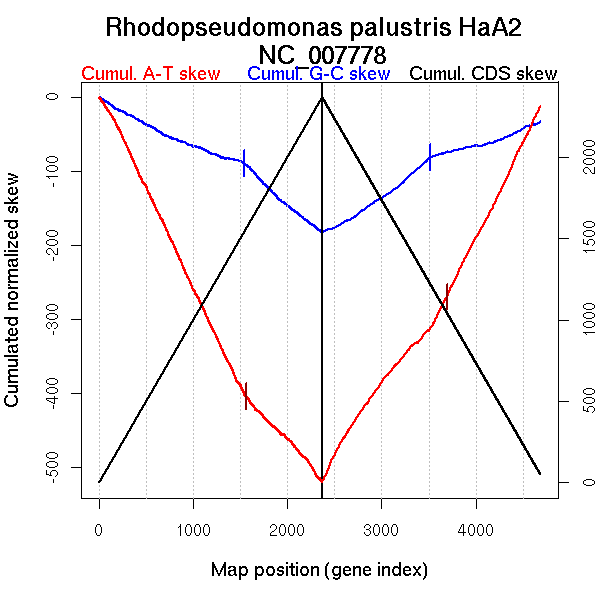
Oriloc predictions: Origin 336 kb Terminus 3095 kb
Worning et al., 2006: Origin 411 kb Terminus 3113 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 2291.361 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 1.38 kb
Consensus predictions: Origin 336 kb Terminus 3095 kb
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 1543 | NA | 3109 |
| GC-skew reverse | 3170 | NA | 1942 |
| 3515 | NA | 3052 | |
| AT-skew forward | 162 | NA | 417 |
| 1458 | NA | 2979 | |
| 1670 | NA | 3491 | |
| 2219 | NA | 5002 | |
| AT-skew reverse | 3694 | NA | 3373 |
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 1543 (3109.323 kb) | leading | -0.058 |
| 1544 (3110.981 kb) | 2368 (5330.976 kb) | lagging | -0.113 | |
| GC-skew reverse | 2369 (0 kb) | 3170 (1941.554 kb) | NA | 0.079 |
| 3171 (1942.3225 kb) | 3515 (3052.103 kb) | leading | 0.115 | |
| 3516 (3054.768 kb) | 4683 (5330.976 kb) | lagging | 0.039 | |
| AT-skew forward | 1 (0 kb) | 162 (416.965 kb) | NA | -0.193 |
| 163 (419.461 kb) | 1458 (2978.7485 kb) | leading | -0.271 | |
| 1459 (2980.167 kb) | 1670 (3490.755 kb) | NA | -0.195 | |
| 1671 (3491.7915 kb) | 2219 (5002.4865 kb) | lagging | -0.125 | |
| 2220 (5014.043 kb) | 2368 (5330.976 kb) | lagging | -0.196 | |
| AT-skew reverse | 2369 (0 kb) | 3694 (3372.6935 kb) | NA | 0.173 |
| 3695 (3374.151 kb) | 4683 (5330.976 kb) | lagging | 0.256 |
More G than C on the leading strand for replication - for forward encoded genes.