Rearranged Oriloc output example: Previous Index Next
Bacteria; Firmicutes; Lactobacillales; Streptococcaceae; Streptococcus.
Genome size (bp): 1838554
Number of genes 1865
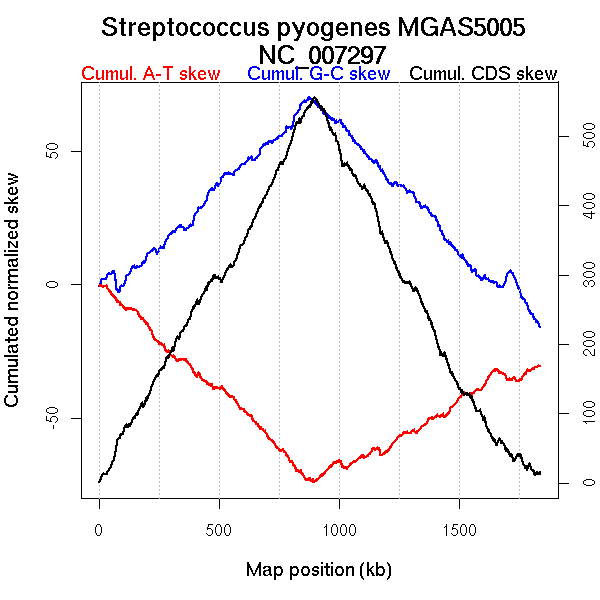
Oriloc predictions: Origin 0 kb Terminus 885 kb
Worning et al., 2006: Origin 1836 kb Terminus 897 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 1757.412 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 0.88 kb
Consensus predictions: Origin 0 kb Terminus 885 kb
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 713 | NA | 875 |
| GC-skew reverse | 1108 | NA | 871 |
| 1769 | NA | 1682 | |
| 1798 | NA | 1718 | |
| AT-skew forward | 123 | NA | 160 |
| 241 | NA | 304 | |
| 292 | NA | 364 | |
| AT-skew reverse | 1054 | NA | 509 |
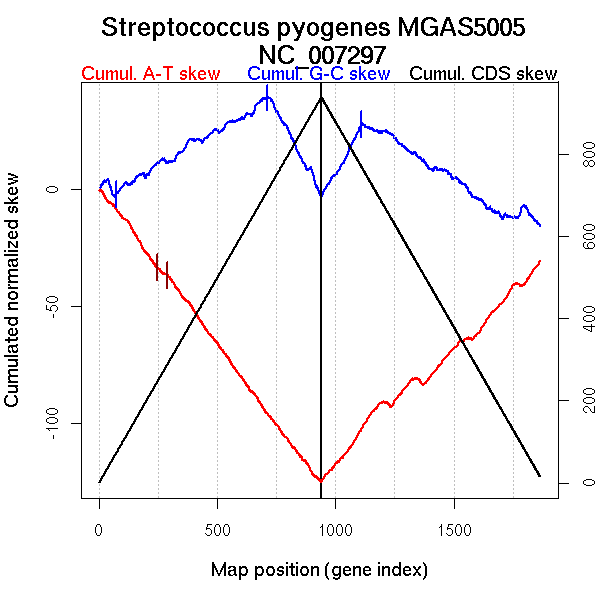
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 713 (875.064 kb) | leading | 0.057 |
| 714 (877.7315 kb) | 940 (1838.52 kb) | lagging | -0.188 | |
| GC-skew reverse | 941 (0 kb) | 1108 (870.9885 kb) | leading | 0.175 |
| 1109 (871.721 kb) | 1769 (1682.2625 kb) | lagging | -0.068 | |
| 1770 (1683.7145 kb) | 1798 (1718.3565 kb) | lagging | 0.231 | |
| 1799 (1720.371 kb) | 1865 (1838.52 kb) | lagging | -0.131 | |
| AT-skew forward | 1 (0 kb) | 123 (160.389 kb) | leading | -0.116 |
| 124 (162.011 kb) | 241 (304.3065 kb) | leading | -0.163 | |
| 242 (304.8705 kb) | 292 (364.1815 kb) | leading | -0.072 | |
| 293 (364.9035 kb) | 940 (1838.52 kb) | NA | -0.138 | |
| AT-skew reverse | 941 (0 kb) | 1054 (509.408 kb) | leading | 0.113 |
| 1055 (510.426 kb) | 1865 (1838.52 kb) | lagging | 0.091 |
More G than C on the leading strand for replication.