Rearranged Oriloc output example: Previous Index Next
Archaea; Euryarchaeota; Halobacteria; Halobacteriales; Halobacteriaceae; Natronomonas.
Genome size (bp): 2595221
Number of genes 2661
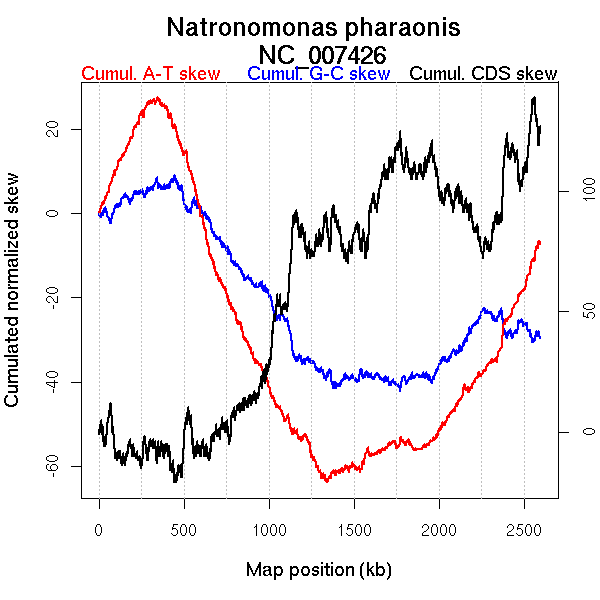
Oriloc predictions: Origin 339 kb Terminus 1320 kb
Worning et al., 2006: Origin 1334 kb Terminus 301 kb
Position(s) of the ORC/Cdc6 gene(s): 304.09 kb, 1488.11 kb, 1567.16 kb, 299.36 kb, 304.09 kb, 1488.11 kb, 1567.16 kb
Consensus predictions: Origin 339 kb Terminus 1320 kb
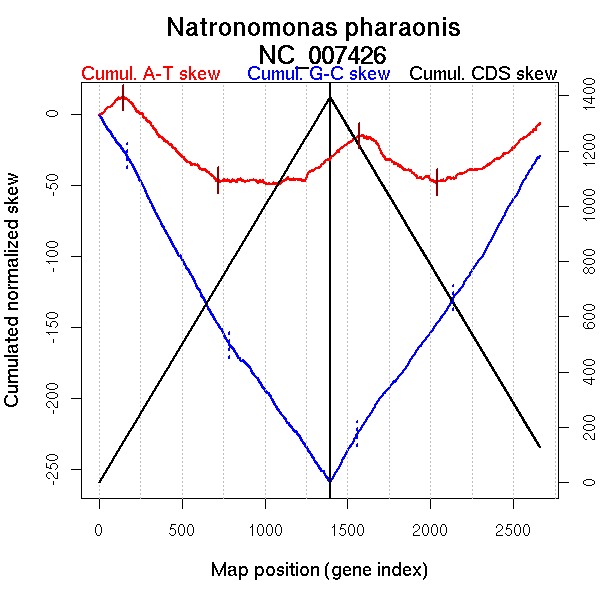
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 168 | 0.06333 | 348 |
| 788 | 0.05333 | 1465 | |
| 168 | 0.06333 | 348 | |
| 788 | 0.05333 | 1465 | |
| GC-skew reverse | 1558 | 0.05333 | 333 |
| 2139 | 0.11667 | 1533 | |
| 1558 | 0.05333 | 333 | |
| 2139 | 0.11667 | 1533 | |
| AT-skew forward | 146 | 0.00333 | 307 |
| 719 | 0.02333 | 1322 | |
| AT-skew reverse | 1572 | 0.01333 | 354 |
| 2039 | 0.02667 | 1343 |
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 168 (347.948 kb) | lagging | -0.168 |
| 169 (350.9265 kb) | 168 (347.948 kb) | leading | 0.21 | |
| 169 (350.9265 kb) | 788 (1465.195 kb) | leading | -0.215 | |
| 789 (1465.921 kb) | 788 (1465.195 kb) | lagging | 0.1 | |
| 789 (1465.921 kb) | 1394 (2595.209 kb) | lagging | -0.161 | |
| GC-skew reverse | 1395 (0 kb) | 1558 (332.7265 kb) | lagging | 0.207 |
| 1559 (334.33 kb) | 1558 (332.7265 kb) | lagging | -0.079 | |
| 1559 (334.33 kb) | 2139 (1533.052 kb) | NA | 0.159 | |
| 2140 (1534.0115 kb) | 2139 (1533.052 kb) | lagging | -0.617 | |
| 2140 (1534.0115 kb) | 2661 (2595.209 kb) | lagging | 0.199 | |
| AT-skew forward | 1 (0 kb) | 146 (306.632 kb) | lagging | 0.093 |
| 147 (308.245 kb) | 719 (1322.08 kb) | leading | -0.106 | |
| 720 (1322.8535 kb) | 1394 (2595.209 kb) | lagging | 0.016 | |
| AT-skew reverse | 1395 (0 kb) | 1572 (354.3655 kb) | lagging | 0.09 |
| 1573 (355.4835 kb) | 2039 (1342.621 kb) | leading | -0.078 | |
| 2040 (1343.576 kb) | 2661 (2595.209 kb) | lagging | 0.064 |
More C than G on the leading strand for replication - for forward encoded genes.