Rearranged Oriloc output example: Previous Index Next
Bacteria; Chlorobi; Chlorobia; Chlorobiales; Chlorobiaceae; Chlorobium/Pelodictyon group; Chlorobium.
Genome size (bp): 2572079
Number of genes 2002
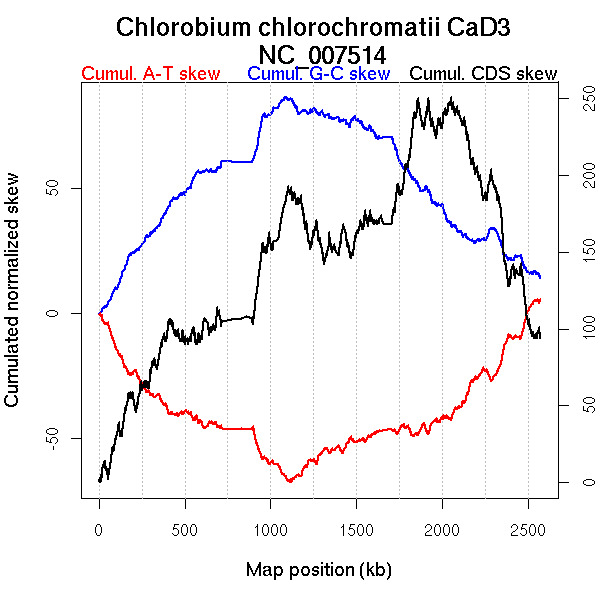
Oriloc predictions: Origin 0 kb Terminus 1118 kb
Worning et al., 2006: Origin 2559 kb Terminus 1110 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 0.059 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 1.08 kb
Consensus predictions: Origin 0 kb Terminus 1118 kb
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 468 | 0 | 1083 |
| GC-skew reverse | 1318 | 0 | 972 |
| AT-skew forward | 484 | 0 | 1114 |
| AT-skew reverse | 1402 | 0 | 1217 |
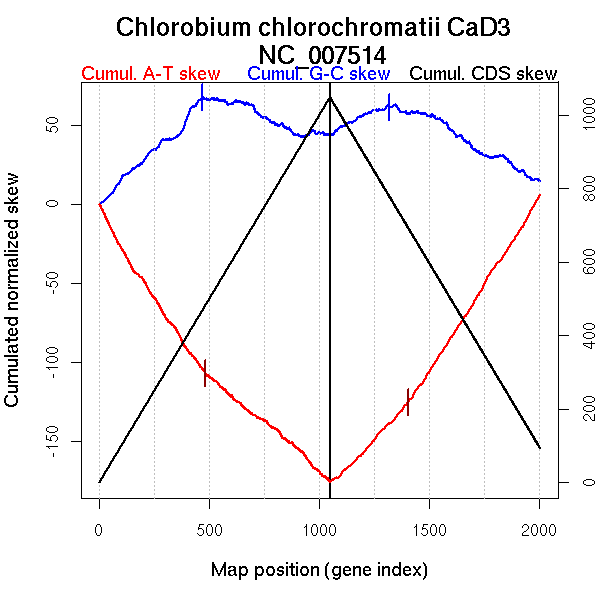
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 468 (1082.9745 kb) | leading | 0.136 |
| 469 (1087.22 kb) | 1048 (2571.844 kb) | lagging | -0.051 | |
| GC-skew reverse | 1049 (0 kb) | 1318 (971.8045 kb) | leading | 0.067 |
| 1319 (973.574 kb) | 2002 (2571.844 kb) | lagging | -0.076 | |
| AT-skew forward | 1 (0 kb) | 484 (1113.6855 kb) | leading | -0.216 |
| 485 (1114.7655 kb) | 1048 (2571.844 kb) | lagging | -0.118 | |
| AT-skew reverse | 1049 (0 kb) | 1402 (1216.655 kb) | NA | 0.145 |
| 1403 (1217.3005 kb) | 2002 (2571.844 kb) | lagging | 0.218 |
More G than C on the leading strand for replication.