Rearranged Oriloc output example: Previous Index Next
Bacteria; Proteobacteria; Alphaproteobacteria; Rhizobiales; Brucellaceae; Brucella.
Genome size (bp): 2121359
Number of genes 2000
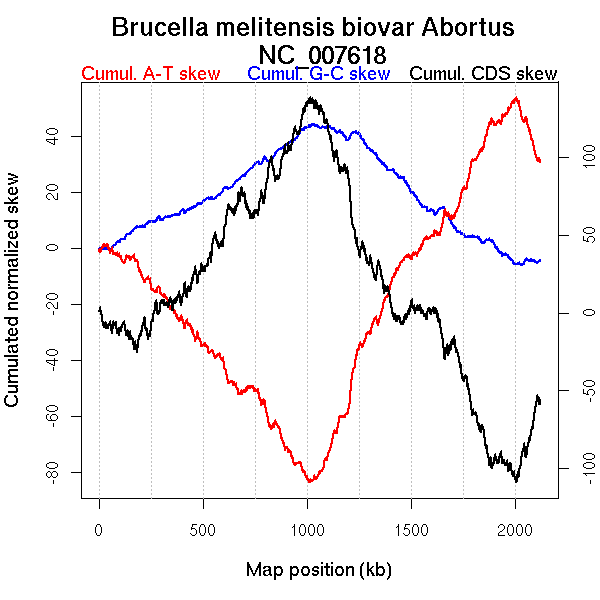
Oriloc predictions: Origin 2007 kb Terminus 1024 kb
Worning et al., 2006: Origin NA kb Terminus NA kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 2007.957 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 1.53 kb, 584.02 kb, 1099.13 kb
Consensus predictions: Origin 2007 kb Terminus 1024 kb
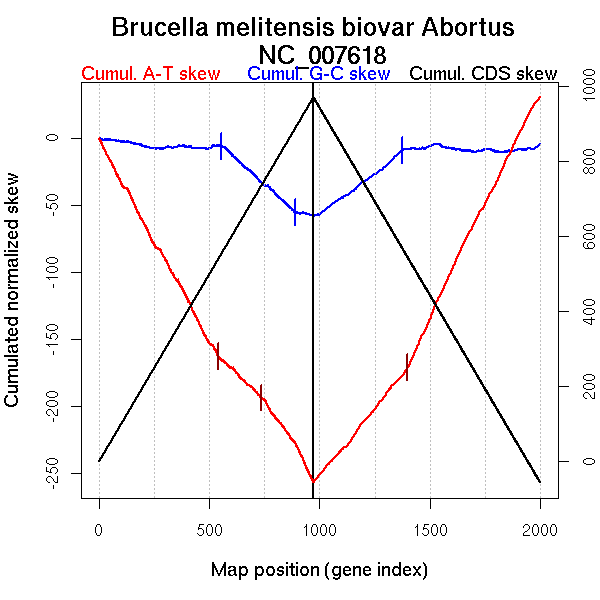
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 555 | NA | 1047 |
| 890 | NA | 2006 | |
| GC-skew reverse | 1373 | NA | 999 |
| AT-skew forward | 542 | NA | 1013 |
| 735 | NA | 1559 | |
| AT-skew reverse | 1399 | NA | 1058 |
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 555 (1046.9695 kb) | leading | -0.011 |
| 556 (1053.557 kb) | 890 (2006.217 kb) | lagging | -0.148 | |
| 891 (2008.926 kb) | 972 (2121.15 kb) | leading | -0.026 | |
| GC-skew reverse | 973 (0 kb) | 1373 (998.8635 kb) | leading | 0.122 |
| 1374 (1002.1865 kb) | 2000 (2121.15 kb) | lagging | -0.003 | |
| AT-skew forward | 1 (0 kb) | 542 (1012.669 kb) | leading | -0.299 |
| 543 (1013.036 kb) | 735 (1559.4045 kb) | lagging | -0.156 | |
| 736 (1561.8305 kb) | 972 (2121.15 kb) | NA | -0.259 | |
| AT-skew reverse | 973 (0 kb) | 1399 (1058.3955 kb) | leading | 0.201 |
| 1400 (1059.286 kb) | 2000 (2121.15 kb) | lagging | 0.339 |
More G than C on the leading strand for replication.