Rearranged Oriloc output example: Previous Index Next
Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Erythrobacteraceae; Erythrobacter.
Genome size (bp): 3052398
Number of genes 3010
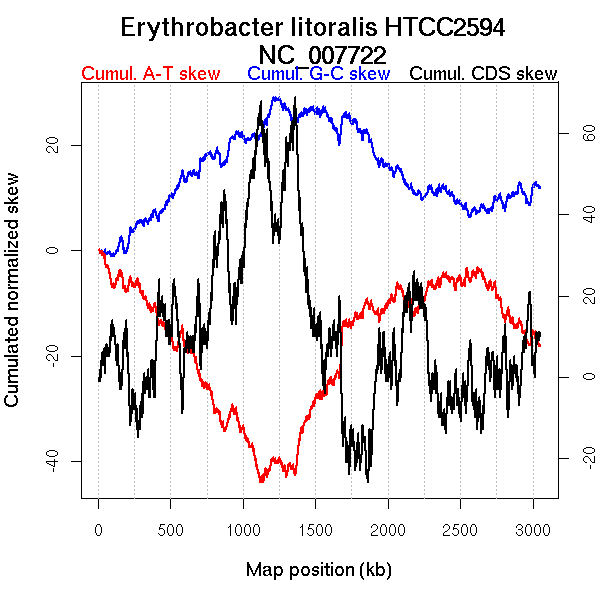
Oriloc predictions: Origin 0 kb Terminus 1207 kb
Worning et al., 2006: Origin 2626 kb Terminus 1298 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 1188.291 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 2143.12 kb
Consensus predictions: Origin 0 kb Terminus 1207 kb
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 607 | 0 | 1218 |
| GC-skew reverse | 2097 | 0.02 | 1251 |
| AT-skew forward | 523 | 0.04 | 1032 |
| AT-skew reverse | 1963 | 0.02667 | 958 |
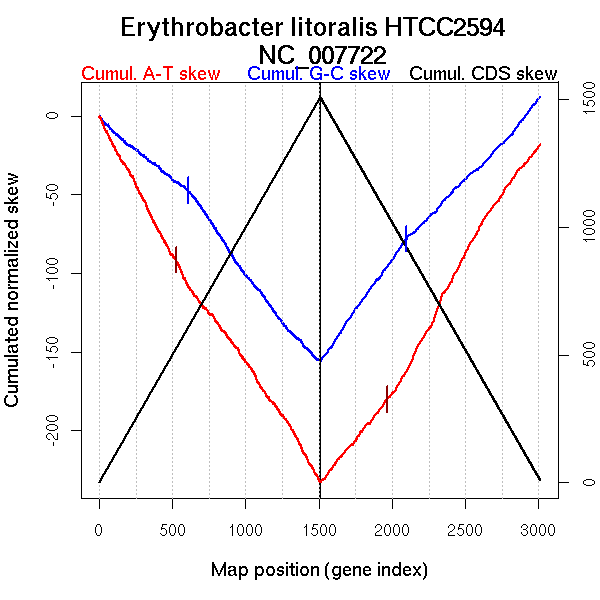
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 607 (1218.236 kb) | leading | -0.075 |
| 608 (1218.7185 kb) | 1510 (3050.991 kb) | lagging | -0.124 | |
| GC-skew reverse | 1511 (0 kb) | 2097 (1250.7195 kb) | leading | 0.133 |
| 2098 (1254.263 kb) | 3010 (3050.991 kb) | lagging | 0.097 | |
| AT-skew forward | 1 (0 kb) | 523 (1032.082 kb) | leading | -0.176 |
| 524 (1035.2365 kb) | 1510 (3050.991 kb) | lagging | -0.136 | |
| AT-skew reverse | 1511 (0 kb) | 1963 (957.807 kb) | leading | 0.115 |
| 1964 (961.255 kb) | 3010 (3050.991 kb) | lagging | 0.164 |
More G than C on the leading strand for replication.