Rearranged Oriloc output example: Previous Index Next
Bacteria; Proteobacteria; Deltaproteobacteria; Syntrophobacterales; Syntrophaceae; Syntrophus.
Genome size (bp): 3179300
Number of genes 3168
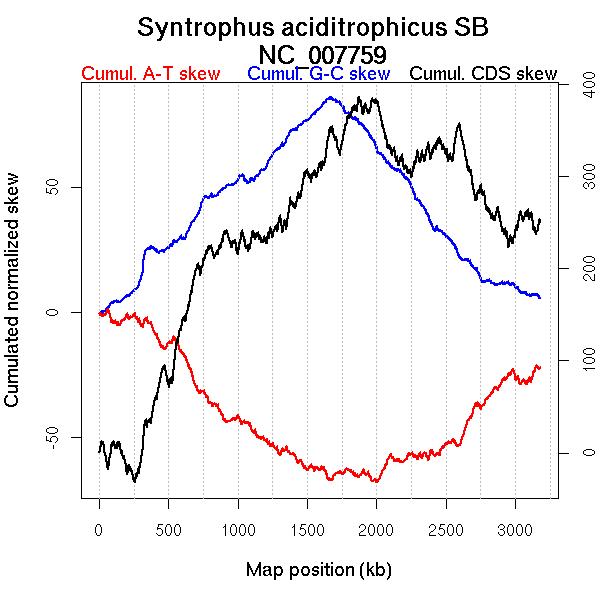
Oriloc predictions: Origin 0 kb Terminus 1665 kb
Worning et al., 2006: Origin 3149 kb Terminus 1798 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 859.145 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 0.84 kb
Consensus predictions: Origin 0 kb Terminus 1665 kb
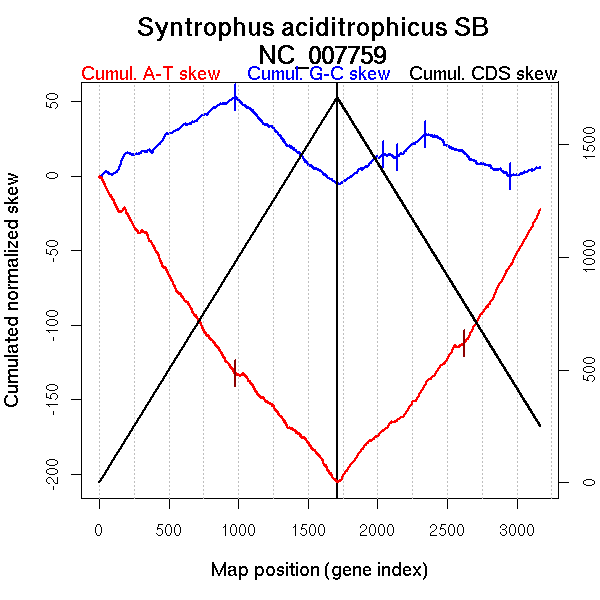
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 980 | 0 | 1660 |
| GC-skew reverse | 2043 | 0.03 | 936 |
| 2141 | 0.02167 | 1123 | |
| 2343 | 0.00667 | 1668 | |
| 2956 | 0.04833 | 2782 | |
| AT-skew forward | 975 | 0.01333 | 1656 |
| AT-skew reverse | 2622 | 0.00667 | 2205 |
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 980 (1660.08 kb) | leading | 0.055 |
| 981 (1661.6295 kb) | 1710 (3178.51 kb) | lagging | -0.082 | |
| GC-skew reverse | 1711 (0 kb) | 2043 (936.269 kb) | leading | 0.065 |
| 2044 (938.773 kb) | 2141 (1122.8715 kb) | leading | -0.024 | |
| 2142 (1124.362 kb) | 2343 (1668.4735 kb) | leading | 0.076 | |
| 2344 (1669.327 kb) | 2956 (2782.393 kb) | lagging | -0.048 | |
| 2957 (2783.173 kb) | 3168 (3178.51 kb) | lagging | 0.029 | |
| AT-skew forward | 1 (0 kb) | 975 (1656.4535 kb) | leading | -0.136 |
| 976 (1657.2495 kb) | 1710 (3178.51 kb) | lagging | -0.101 | |
| AT-skew reverse | 1711 (0 kb) | 2622 (2205.2075 kb) | NA | 0.102 |
| 2623 (2205.943 kb) | 3168 (3178.51 kb) | lagging | 0.163 |
More G than C on the leading strand for replication.