Rearranged Oriloc output example: Previous Index Next
Archaea; Euryarchaeota; Methanomicrobia; Methanomicrobiales; Methanospirillaceae; Methanospirillum.
Genome size (bp): 3544738
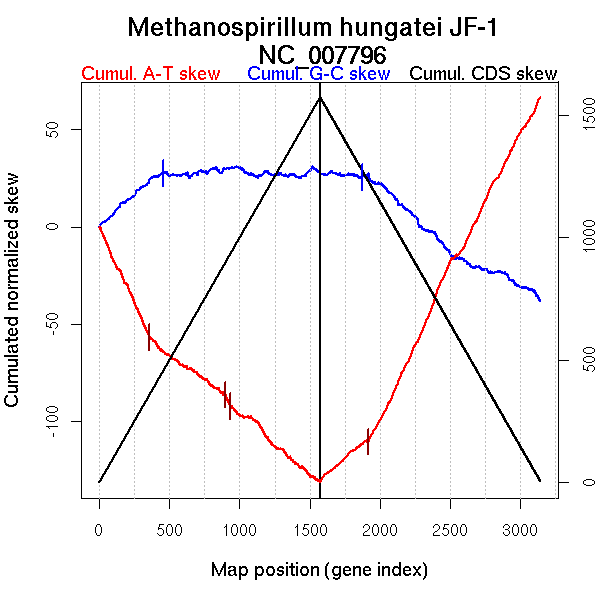
Number of genes 3139
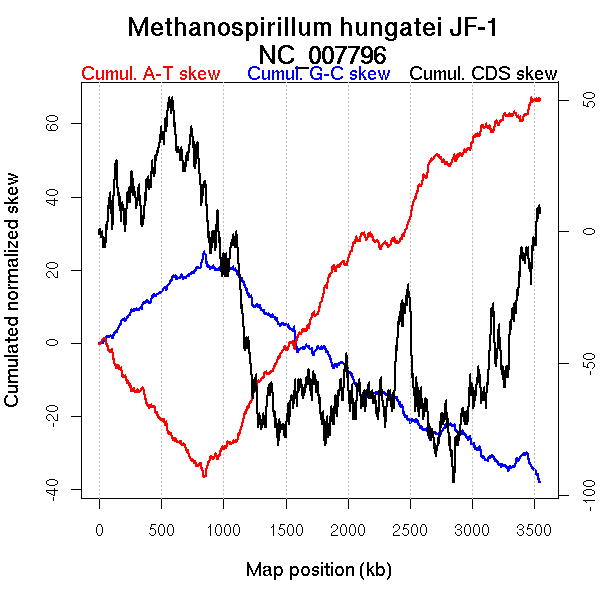
Oriloc predictions: Origin 0 kb Terminus 844 kb
Worning et al., 2006: Origin 3494 kb Terminus 1022 kb
Position(s) of the ORC/Cdc6 gene(s): 518.2 kb
Consensus predictions: Origin 0 kb Terminus 844 kb
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 454 | 0 | 1052 |
| GC-skew reverse | 1870 | 0.02 | 759 |
| AT-skew forward | 359 | 0 | 838 |
| 900 | 0 | 2176 | |
| 936 | 0.00222 | 2259 | |
| AT-skew reverse | 1918 | 0 | 849 |
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 454 (1052.0045 kb) | NA | 0.063 |
| 455 (1053.521 kb) | 1573 (3544.709 kb) | lagging | 0 | |
| GC-skew reverse | 1574 (0 kb) | 1870 (758.7745 kb) | leading | -0.005 |
| 1871 (760.5225 kb) | 3139 (3544.709 kb) | lagging | -0.051 | |
| AT-skew forward | 1 (0 kb) | 359 (838.3355 kb) | leading | -0.156 |
| 360 (841.5225 kb) | 900 (2175.9855 kb) | lagging | -0.052 | |
| 901 (2177.611 kb) | 936 (2259.1985 kb) | lagging | -0.157 | |
| 937 (2260.5035 kb) | 1573 (3544.709 kb) | lagging | -0.065 | |
| AT-skew reverse | 1574 (0 kb) | 1918 (849.12 kb) | leading | 0.062 |
| 1919 (854.457 kb) | 3139 (3544.709 kb) | lagging | 0.147 |
More G than C on the leading strand for replication - for reverse encoded genes.