Defines the basic types of data flow nodes. More...
Namespaces | |
| namespace | BrentOneDimension |
| namespace | Graph |
| namespace | IntegerTools |
| namespace | NewtonBacktrackOneDimension |
| namespace | numeric |
| namespace | PowellMultiDimensions |
| namespace | StatTools |
| namespace | TextTools |
| namespace | VectorTools |
Classes | |
| class | AAChargeIndex |
| class | AAChenGuHuangHydrophobicityIndex |
| class | AAChouFasmanAHelixIndex |
| class | AAChouFasmanBSheetIndex |
| class | AAChouFasmanTurnIndex |
| class | AAExteriorSubstitutionRegister |
| Indexes only substitutions between amino-acids. Groups are distinguished by their direction. More... | |
| class | AAIndex1Entry |
| class | AAIndex2Entry |
| class | AAInteriorSubstitutionRegister |
| Indexes only intra amino-acid substitutions. Every group represents a substitutions for the same amino acid. Met and Trp are not taken into account due their non-degenerescence. More... | |
| class | AAMassIndex |
| class | AASEA1030Index |
| class | AASEAInf10Index |
| class | AASEASup30Index |
| class | AASurfaceIndex |
| class | AAVolumeIndex |
| class | AbstractAgglomerativeDistanceMethod |
| Partial implementation of the AgglomerativeDistanceMethod interface. More... | |
| class | AbstractAlignedPhyloLikelihood |
| class | AbstractAlignedPhyloLikelihoodSet |
| The AlignedPhyloLikelihoodSet abstract class. More... | |
| class | AbstractAlphabet |
| class | AbstractAutonomousSubstitutionProcess |
| A partial implementation of the SubstitutionProcess interface. More... | |
| class | AbstractBiblioMixedTransitionModel |
| Abstract class for mixture models based on the bibliography. More... | |
| class | AbstractBiblioSubstitutionModel |
| class | AbstractBiblioTransitionModel |
| Partial implementation of the SubstitutionModel interface for models that are set for matching the bibliography, and are only defined through a link to a "real" model. More... | |
| class | AbstractCodonAAFitnessSubstitutionModel |
| Abstract class for modelling of ratios of substitution rates between codons, whatever they are synonymous or not. More... | |
| class | AbstractCodonAARateSubstitutionModel |
| Abstract class for modelling of non-synonymous and synonymous substitution rates in codon models, given an amino acid rate matrix (from a shared_ptr model). More... | |
| class | AbstractCodonBGCSubstitutionModel |
| Abstract class for modelling of non-synonymous and synonymous substitution rates in codon models, with gBGC. More... | |
| class | AbstractCodonClusterAASubstitutionModel |
| Abstract class for modelling of non-synonymous and synonymous substitution rates in codon models, with AA clustered. More... | |
| class | AbstractCodonCpGSubstitutionModel |
| Abstract class for modelling of CpG -> CpA or TpG (symmetric) hypermutability substitution rate inside codons. Note that the neihbouring effects between codons are not considered. More... | |
| class | AbstractCodonDistanceSubstitutionModel |
| Abstract class for modelling of non-synonymous and synonymous substitution rates in codon models. More... | |
| class | AbstractCodonFitnessSubstitutionModel |
| Abstract class for modelling of ratios of substitution rates between codons, whatever they are synonymous or not. More... | |
| class | AbstractCodonFrequenciesSubstitutionModel |
| Abstract Class for substitution models on codons parametrized by frequencies. More... | |
| class | AbstractCodonPhaseFrequenciesSubstitutionModel |
| Abstract Class for substitution models on codons parametrized by a frequency. More... | |
| class | AbstractCodonSubstitutionModel |
| Abstract class for substitution models on codons. More... | |
| class | AbstractColorSet |
| class | AbstractCoreSequence |
| class | AbstractCoreSite |
| class | AbstractDendrogramPlot |
| Basic implementation of dendrogram plots. More... | |
| class | AbstractDFPSubstitutionModel |
| Class for neutral substitution models on triplets, following the mutation process proposed in Doron-Fagenboim & Pupko, 2006, but without equilibrium frequencies. This model is an extension of Kimura 2-rates substitution model to codons. More... | |
| class | AbstractDiscreteDistribution |
| class | AbstractDiscreteRatesAcrossSitesTreeLikelihood |
| Partial implementation of the DiscreteRatesAcrossSitesTreeLikelihood interface. More... | |
| class | AbstractFontManager |
| class | AbstractFrequencySet |
| Basic implementation of the FrequencySet interface. More... | |
| class | AbstractFromSubstitutionModelTransitionModel |
| Virtual class of a Transition Model related to a given SubstitutionModel. More... | |
| class | AbstractGraphicDevice |
| class | AbstractHmmLikelihood |
| class | AbstractHmmTransitionMatrix |
| class | AbstractHomogeneousTreeLikelihood |
| Partial implementation for homogeneous model of the TreeLikelihood interface. More... | |
| class | AbstractIAlignment |
| class | AbstractIAlignment2 |
| class | AbstractIDistanceMatrix |
| class | AbstractIMultiPhyloDAG |
| Partial implementation of the IMultiDAG interface. More... | |
| class | AbstractIMultiPhyloTree |
| Partial implementation of the IMultiTree interface. More... | |
| class | AbstractIMultiTree |
| Partial implementation of the IMultiTree interface. More... | |
| class | AbstractIPhyloDAG |
| Partial implementation of the IDAG interface. More... | |
| class | AbstractIPhyloTree |
| class | AbstractIProbabilisticAlignment |
| class | AbstractIProbabilisticAlignment2 |
| class | AbstractIProbabilisticSequence |
| class | AbstractISequence |
| class | AbstractITree |
| Partial implementation of the ITree interface. More... | |
| class | AbstractKroneckerCodonSubstitutionModel |
| Abstract class for substitution models on codons allowing multiple substitutions. More... | |
| class | AbstractKroneckerWordSubstitutionModel |
| Abstract Kronecker Word Model. More... | |
| class | AbstractLkTransitionModel |
| Partial implementation of the TransitionModel interface, with function for likelihood computations. More... | |
| class | AbstractMapping |
| Partial implementation of the mapping interface. More... | |
| class | AbstractMixedTransitionModel |
| Partial implementation for Mixed Transition models, defined as a mixture of "simple" substitution models. Each model has a specific probability and rate, with the constraint that the expectation (on the distribution of the models) of the rate of all the models equals one. More... | |
| class | AbstractMutationProcess |
| Partial implementation of the MutationProcess interface. More... | |
| class | AbstractNonHomogeneousTreeLikelihood |
| Partial implementation for branch non-homogeneous models of the TreeLikelihood interface. More... | |
| class | AbstractNucleotideSubstitutionModel |
| Specialisation abstract class for nucleotide substitution model. More... | |
| class | AbstractNumericalDerivative |
| class | AbstractOAlignment |
| class | AbstractODistanceMatrix |
| class | AbstractOMultiPhyloDAG |
| Partial implementation of the ODAG interface. More... | |
| class | AbstractOMultiPhyloTree |
| Partial implementation of the OTree interface. More... | |
| class | AbstractOMultiTree |
| Partial implementation of the OTree interface. More... | |
| class | AbstractOPhyloDAG |
| Partial implementation of the ODAG interface. More... | |
| class | AbstractOPhyloTree |
| Partial implementation of the OTree interface. More... | |
| class | AbstractOProbabilisticAlignment |
| class | AbstractOProbabilisticSequence |
| class | AbstractOProbabilisticSequence2 |
| class | AbstractOptimizationStopCondition |
| class | AbstractOptimizer |
| class | AbstractOSequence |
| class | AbstractOSequence2 |
| class | AbstractOTree |
| Partial implementation of the OTree interface. More... | |
| class | AbstractOutputStream |
| class | AbstractParameterAliasable |
| class | AbstractParametrizable |
| class | AbstractParametrizableSequencePhyloLikelihood |
| class | AbstractPhyloLikelihood |
| class | AbstractPhyloLikelihoodSet |
| The PhyloLikelihoodSet class, to manage a subset of PhyloLikelihoods from a given PhyloLikelihoodContainer. More... | |
| class | AbstractProteinSubstitutionModel |
| Specialisation abstract class for protein substitution model. More... | |
| class | AbstractReverseTransliterator |
| class | AbstractReversibleNucleotideSubstitutionModel |
| Specialisation abstract class for reversible nucleotide substitution model. More... | |
| class | AbstractReversibleProteinSubstitutionModel |
| Specialisation abstract class for reversible protein substitution model. More... | |
| class | AbstractReversibleSubstitutionModel |
| Partial implementation of the ReversibleSubstitutionModel interface. More... | |
| class | AbstractReward |
| Basic implementation of the the Reward interface. More... | |
| class | AbstractRewardMapping |
| Partial implementation of the substitution mapping interface. More... | |
| class | AbstractSequencePhyloLikelihood |
| class | AbstractSequencePositionIterator |
| class | AbstractSingleDataPhyloLikelihood |
| class | AbstractSinglePhyloSubstitutionMapping |
| The AbstractSinglePhyloSubstitutionMapping class: substitution mapping linked with a Single Process PhyloLikelihood. More... | |
| class | AbstractStateMap |
| A convenience partial implementation of the StateMap interface. More... | |
| class | AbstractSubstitutionCount |
| Partial implementation of the SubstitutionCount interface. More... | |
| class | AbstractSubstitutionDistance |
| Partial implementation of the SubstitutionDistance interface. More... | |
| class | AbstractSubstitutionMapping |
| Partial implementation of the substitution mapping interface. More... | |
| class | AbstractSubstitutionModel |
| class | AbstractSubstitutionProcess |
| A partial implementation of the SubstitutionProcess interface. More... | |
| class | AbstractSubstitutionRegister |
| class | AbstractTemplateEventDrivenSymbolList |
| class | AbstractTemplateSequenceContainer |
| class | AbstractTemplateSiteContainerIterator |
| class | AbstractTemplateSymbolList |
| class | AbstractTotallyWrappedSubstitutionModel |
| class | AbstractTotallyWrappedTransitionModel |
| class | AbstractTransitionModel |
| Partial implementation of the TransitionModel interface. More... | |
| class | AbstractTransliterator |
| class | AbstractTreeDrawing |
| Partial implementation of the TreeDrawing interface. More... | |
| class | AbstractTreeLikelihood |
| Partial implementation of the TreeLikelihood interface. More... | |
| class | AbstractTreeLikelihoodData |
| Partial implementation of the TreeLikelihoodData interface. More... | |
| class | AbstractTreeParsimonyData |
| Partial implementation of the TreeParsimonyData interface. More... | |
| class | AbstractTreeParsimonyScore |
| Partial implementation of the TreeParsimonyScore interface. More... | |
| class | AbstractWeightedSubstitutionCount |
| Partial implementation of the WeightedSubstitutionCount interface. More... | |
| class | AbstractWordFrequencySet |
| class | AbstractWordSubstitutionModel |
| Abstract Basal class for words of substitution models. More... | |
| class | AbstractWrappedModel |
| Abstract class of Wrapping model class, where all methods are redirected from model(). More... | |
| class | AbstractWrappedSubstitutionModel |
| class | AbstractWrappedTransitionModel |
| class | AdaptiveKernelDensityEstimation |
| class | AgglomerativeDistanceMethodInterface |
| Interface for agglomerative distance methods. More... | |
| class | AliasParameterListener |
| class | AlignedLikelihoodCalculation |
| class | AlignedPhyloLikelihoodAutoCorrelation |
| Likelihood framework based on a hmm of simple likelihoods. More... | |
| class | AlignedPhyloLikelihoodHmm |
| Likelihood framework based on a hmm of simple likelihoods. More... | |
| class | AlignedPhyloLikelihoodInterface |
| The AlignedPhyloLikelihood interface, for phylogenetic likelihood. More... | |
| class | AlignedPhyloLikelihoodMixture |
| Likelihood framework based on a mixture of aligned likelihoods. More... | |
| class | AlignedPhyloLikelihoodProduct |
| The AlignedPhyloLikelihoodProduct class, for phylogenetic likelihood on several independent data. More... | |
| class | AlignedPhyloLikelihoodSetInterface |
| Joint interface SetOf+Aligned PhylLikelihood. More... | |
| class | AllelicAlphabet |
| class | Alphabet |
| class | AlphabetException |
| class | AlphabetIndex1 |
| class | AlphabetIndex2 |
| class | AlphabetMismatchException |
| class | AlphabetNumericState |
| class | AlphabetState |
| class | AlphabetTools |
| class | AncestralStateReconstruction |
| Interface for ancestral states reconstruction methods. More... | |
| class | AnonymousSubstitutionModel |
| class | ApplicationTools |
| class | AscidianMitochondrialGeneticCode |
| class | AssociationDAGraphImplObserver |
| class | AssociationDAGraphObserver |
| class | AssociationGraphImplObserver |
| class | AssociationGraphObserver |
| class | AssociationTreeGraphImplObserver |
| class | AssociationTreeGraphObserver |
| class | AttributesTools |
| class | AutoCorrelationProcessPhyloLikelihood |
| Likelihood framework based on an auto-correlation of simple likelihoods. More... | |
| class | AutoCorrelationSequenceEvolution |
| Sequence evolution framework based on an auto-correlation of substitution processes. More... | |
| class | AutoCorrelationTransitionMatrix |
| class | AutonomousSubstitutionProcessInterface |
| Interface for SubstitutionProcess objects that own their own ParametrizablePhyloTree & Scenario. More... | |
| class | AutoParameter |
| class | AwareNode |
| A node class aware of its neighbours. More... | |
| class | BackupListener |
| class | BackwardHmmLikelihood_DF |
| class | BackwardLikelihoodTree |
| class | BadCharException |
| class | BadIntegerException |
| class | BadIntException |
| class | BadNumberException |
| class | BadSizeException |
| class | BasicTreeDrawingDisplayControler |
| Easy tune of tree drawings display, a basic implementation: More... | |
| class | BetaDiscreteDistribution |
| class | BfgsMultiDimensions |
| class | BinaryAlphabet |
| class | BinaryOperator |
| class | BinarySubstitutionModel |
| The Model on two states. More... | |
| class | BioNJ |
| The BioNJ distance method. More... | |
| class | BipartitionList |
| This class deals with the bipartitions defined by trees. More... | |
| class | BipartitionTools |
| This class provides tools related to the BipartitionList class. More... | |
| class | BLOSUM50 |
| class | BootstrapValuesTreeDrawingListener |
| A TreeDrawingListener implementation that write the bootstrap values of inner nodes. More... | |
| class | BowkerTest |
| class | BppApplication |
| class | BppBoolean |
| class | BppDouble |
| class | BppInteger |
| class | BppNotANumber |
| class | BppNumberI |
| class | BppOAlignmentReaderFormat |
| class | BppOAlignmentWriterFormat |
| class | BppOAlphabetIndex1Format |
| class | BppOAlphabetIndex2Format |
| class | BppOBranchModelFormat |
| Branch model I/O in BppO format. More... | |
| class | BppODiscreteDistributionFormat |
| class | BppOFrequencySetFormat |
| Frequencies set I/O in BppO format. More... | |
| class | BppOMultiTreeReaderFormat |
| Tree I/O in BppO format. More... | |
| class | BppOMultiTreeWriterFormat |
| Tree I/O in BppO format. More... | |
| class | BppOParametrizableFormat |
| class | BppORateDistributionFormat |
| Rate Distribution I/O in BppO format. More... | |
| class | BppOSequenceReaderFormat |
| class | BppOSequenceStreamReaderFormat |
| class | BppOSequenceWriterFormat |
| class | BppOSubstitutionModelFormat |
| Substitution model I/O in BppO format. More... | |
| class | BppOTransitionModelFormat |
| Transition model I/O in BppO format. More... | |
| class | BppOTreeReaderFormat |
| Tree I/O in BppO format. More... | |
| class | BppOTreeWriterFormat |
| Tree I/O in BppO format. More... | |
| class | BppPhylogeneticsApplication |
| class | BppSequenceApplication |
| class | BppString |
| class | BppUnsignedInteger |
| class | BppVector |
| class | Bracket |
| class | BracketPoint |
| class | BranchedModelSet |
| class | BranchLengthsTreeDrawingListener |
| A TreeDrawingListener implementation that write the branch lengths of inner nodes. More... | |
| class | BranchLikelihood |
| Compute likelihood for a 4-tree. More... | |
| class | BranchModelInterface |
| Interface for all Branch models. More... | |
| class | BrentOneDimension |
| class | CanonicalStateMap |
| This class implements a state map where all resolved states are modeled. More... | |
| class | CaseMaskedAlphabet |
| class | CategoryFromDiscreteDistribution |
| class | CategorySubstitutionRegister |
| The CategorySubstitutionRegisters. More... | |
| class | CharStateNotSupportedException |
| class | CiliateNuclearGeneticCode |
| class | CladogramDrawBranchEvent |
| class | CladogramPlot |
| Cladogram representation of trees. More... | |
| class | Clonable |
| class | Clustal |
| class | ClusterInfos |
| class | Coala |
| The Coala branch-heterogeneous amino-acid substitution model. More... | |
| class | CoalaCore |
| This class is the core class inherited by the Coala class. COaLA is a branch-heterogeneous amino-acid substitution model. More... | |
| class | CodonAdHocSubstitutionModel |
| Class for substitution models of codons with several layers of codon models. More... | |
| class | CodonAlphabet |
| class | CodonDistanceFrequenciesSubstitutionModel |
| Class for asynonymous substitution models on codons with parameterized equilibrium frequencies and nucleotidic models. More... | |
| class | CodonDistancePhaseFrequenciesSubstitutionModel |
| Class for asynonymous substitution models on codons with parameterized equilibrium frequencies and nucleotidic basic models. More... | |
| class | CodonDistanceSubstitutionModel |
| Class for substitution models of codons with non-synonymous/synonymous ratios of substitution rates defined through a distance between amino-acids. More... | |
| class | CodonFrequencySetInterface |
| Parametrize a set of state frequencies for codons. More... | |
| class | CodonFromIndependentFrequencySet |
| the Frequencies in codons are the product of Independent Frequencies in letters with the frequencies of stop codons set to zero. More... | |
| class | CodonFromProteicAlphabetIndex1 |
| class | CodonFromProteicAlphabetIndex2 |
| class | CodonFromUniqueFrequencySet |
| the Frequencies in codons are the product of the frequencies for a unique FrequencySet in letters, with the frequencies of More... | |
| class | CodonReversibleSubstitutionModelInterface |
| Interface for reversible codon models. More... | |
| class | CodonSameAARateSubstitutionModel |
| Class for modelling of non-synonymous rates in codon models, such that the substitution rates between amino acids are similar to the ones in an amino acid rate matrix (from a shared_ptr model). More... | |
| class | CodonSiteTools |
| class | CodonSubstitutionModelInterface |
| class | CollectionNodes |
| class | ColMatrix |
| class | ColorManager |
| class | ColorSet |
| class | ColorTools |
| class | CombineDeltaShifted |
| class | Commentable |
| class | CompleteSubstitutionRegister |
| Completion of a given substitution register to consider all substitutions. The new substitutions are considered in an additional type. More... | |
| class | CompleteTemplateSiteContainerIterator |
| class | ComprehensiveSubstitutionRegister |
| Distinguishes all types of substitutions. More... | |
| class | CompressedVectorSiteContainer |
| class | ComputationTree |
| class | CondLikelihood |
| class | ConfiguredDistribution |
| Data flow node representing a DiscreteDistribution configured with parameter values. More... | |
| class | ConfiguredFrequencySet |
| Data flow node representing a Frequencies Set configured with parameter values. More... | |
| class | ConfiguredModel |
| Likelihood transition model. More... | |
| class | ConfiguredParameter |
| Data flow node representing a parameter. More... | |
| class | ConfiguredParametrizable |
| class | ConfiguredSimplex |
| Data flow node representing a Frequencies Set configured with parameter values. More... | |
| class | ConfiguredTransitionMatrix |
| Data flow node representing a TransitionMatrix configured with parameter values. More... | |
| class | ConjugateGradientMultiDimensions |
| class | ConstantDistribution |
| class | ConstantOne |
| r = 1 for each component. More... | |
| class | ConstantOperator |
| class | ConstantRateDistribution |
| class | ConstantTransitionFunction |
| class | ConstantZero |
| r = 0 for each component. More... | |
| class | ConstraintException |
| class | ConstraintInterface |
| class | Context |
| Context for dataflow node construction. More... | |
| class | ContingencyTableGenerator |
| class | ContingencyTableTest |
| class | Convert |
| r = convert(f). More... | |
| class | CoordsTools |
| class | CoreCodonSubstitutionModelInterface |
| Interface for codon models. More... | |
| class | CoreSequenceInterface |
| class | CoreSiteInterface |
| class | CoreSymbolListDeletionEvent |
| class | CoreSymbolListEditionEvent |
| class | CoreSymbolListInsertionEvent |
| class | CoreSymbolListListener |
| class | CoreSymbolListSubstitutionEvent |
| class | CoreWordAlphabet |
| class | CorrespondenceAnalysis |
| class | CruxSymbolListInterface |
| class | Cursor |
| Data structure describing a plotting direction. More... | |
| class | CWiseAdd |
| class | CWiseAdd< R, ReductionOf< T > > |
| class | CWiseAdd< R, std::tuple< T0, T1 > > |
| class | CWiseApply |
| class | CWiseCompound |
| class | CWiseCompound< R, ReductionOf< T > > |
| class | CWiseConstantPow |
| class | CWiseDiv |
| class | CWiseDiv< R, std::tuple< T0, T1 > > |
| class | CWiseExp |
| class | CWiseFill |
| class | CWiseInverse |
| class | CWiseLog |
| class | CWiseMatching |
| class | CWiseMatching< R, ReductionOf< T > > |
| class | CWiseMean |
| class | CWiseMean< R, ReductionOf< T >, P > |
| class | CWiseMean< R, ReductionOf< T >, ReductionOf< P > > |
| class | CWiseMul |
| class | CWiseMul< R, ReductionOf< T > > |
| class | CWiseMul< R, std::tuple< T0, T1 > > |
| class | CWiseNegate |
| class | CWisePattern |
| class | CWiseSub |
| class | CWiseSub< R, std::tuple< T0, T1 > > |
| class | D1WalkSubstitutionModel |
| The D1Walk substitution model for Integer alphabet [0;N-1]. In this model, substitutions are possible between adjacent states only. The model is defined through an equilibrium distribution, and rates towards a state are proportional to its frequency at equilibrium. More... | |
| class | DAGraph |
| class | DAGraphImpl |
| class | DataTable |
| class | DCSE |
| class | DecompositionMethods |
| Methods useful for analytical substitution count and rewards using the eigen decomposition method. More... | |
| class | DecompositionReward |
| Analytical reward using the eigen decomposition method. More... | |
| class | DecompositionSubstitutionCount |
| Analytical substitution count using the eigen decomposition method. More... | |
| class | DefaultAlphabet |
| class | DefaultColorSet |
| class | DefaultNucleotideScore |
| class | DetailedSiteSimulatorInterface |
| This interface adds the dSimulate method to the SiteSimulator interface. More... | |
| class | DFP07 |
| Class for non-synonymous substitution models on codons with parameterized equilibrium frequencies and nucleotidic models, with allowed multiple substitutions as parameterized in DFP model, with correction to mimic AA substitution rates from a given protein substitution model. More... | |
| class | DFPDistanceFrequenciesSubstitutionModel |
| Class for non-synonymous substitution models on codons with parameterized equilibrium frequencies and nucleotidic models, with allowed multiple substitutions as parameterized in DFP model. More... | |
| struct | Dimension |
| Store a dimension for type T. More... | |
| struct | Dimension< char > |
| struct | Dimension< double > |
| Specialisation of Dimension<T> for floating point types. More... | |
| struct | Dimension< Eigen::Matrix< T, Rows, Cols > > |
| Specialisation of Dimension<T> for eigen matrix types. More... | |
| struct | Dimension< ExtendedFloat > |
| struct | Dimension< ExtendedFloatEigen< R, C, EigenType > > |
| struct | Dimension< float > |
| struct | Dimension< Parameter > |
| struct | Dimension< size_t > |
| struct | Dimension< std::string > |
| struct | Dimension< TransitionFunction > |
| struct | Dimension< uint > |
| class | DimensionException |
| class | DirectionFunction |
| class | DirichletDiscreteDistribution |
| class | DiscreteDistributionInterface |
| class | DiscreteRatesAcrossSitesTreeLikelihoodInterface |
| Interface for rate across sites (RAS) implementation. More... | |
| class | DistanceEstimation |
| class | DistanceMatrix |
| class | DistanceMethodInterface |
| General interface for distance-based phylogenetic reconstruction methods. More... | |
| class | DNA |
| class | DNAToRNA |
| class | DnDsSubstitutionRegister |
| Distinguishes synonymous from non-synonymous substitutions. More... | |
| class | DownhillSimplexMethod |
| class | DRASDRTreeLikelihoodData |
| Likelihood data structure for rate across sites models, using a double-recursive algorithm. More... | |
| class | DRASDRTreeLikelihoodLeafData |
| Likelihood data structure for a leaf. More... | |
| class | DRASDRTreeLikelihoodNodeData |
| Likelihood data structure for a node. More... | |
| class | DRASRTreeLikelihoodData |
| discrete Rate Across Sites, (simple) Recursive likelihood data structure. More... | |
| class | DRASRTreeLikelihoodNodeData |
| Likelihood data structure for a node. More... | |
| class | DrawBranchEvent |
| Event class used by TreeDrawing classes. More... | |
| class | DrawIBranchEvent |
| Event class that uses INode object (more efficient than relying on nodes id, but less generic). More... | |
| class | DrawINodeEvent |
| Event class that uses INode object (more efficient than relying on nodes id, but less generic). More... | |
| class | DrawNodeEvent |
| Event class used by TreeDrawing classes. More... | |
| class | DrawTreeEvent |
| Event class used by TreeDrawing classes. More... | |
| class | DRHomogeneousMixedTreeLikelihood |
| A class to compute the average of several DRHomogeneousTreeLikelihood defined from a Mixed Substitution Model. More... | |
| class | DRHomogeneousTreeLikelihood |
| This class implements the likelihood computation for a tree using the double-recursive algorithm. More... | |
| class | DRNonHomogeneousTreeLikelihood |
| This class implements the likelihood computation for a tree using the double-recursive algorithm, allowing for non-homogeneous models of substitutions. More... | |
| class | DRTreeLikelihoodInterface |
| Interface for double-recursive (DR) implementation of the likelihood computation. More... | |
| class | DRTreeLikelihoodTools |
| Utilitary methods dealing with objects implementing the DRTreeLikelihood interface. More... | |
| class | DRTreeParsimonyData |
| Parsimony data structure for double-recursive (DR) algorithm. More... | |
| class | DRTreeParsimonyLeafData |
| Parsimony data structure for a leaf. More... | |
| class | DRTreeParsimonyNodeData |
| Parsimony data structure for a node. More... | |
| class | DRTreeParsimonyScore |
| Double recursive implementation of interface TreeParsimonyScore. More... | |
| class | DSO78 |
| The Dayhoff, Schwartz and Orcutt substitution model for proteins. More... | |
| class | DualityDiagram |
| class | DuplicatedTableColumnNameException |
| class | DuplicatedTableRowNameException |
| class | DvipsColorSet |
| class | EchinodermMitochondrialGeneticCode |
| class | EdgesIteratorClass |
| class | EdgesIteratorClass< Graph::ALLGRAPHITER, is_const > |
| class | EdgesIteratorClass< Graph::INCOMINGNEIGHBORITER, is_const > |
| class | EdgesIteratorClass< Graph::OUTGOINGNEIGHBORITER, is_const > |
| class | EigenValue |
| class | ElementNotFoundException |
| class | EmptyContainerException |
| class | EmptySequenceException |
| class | EmptySiteException |
| class | EmptyVectorException |
| class | EquilibriumFrequenciesFromModel |
| class | EquilibriumFrequenciesFromTransitionMatrix |
| class | EquiprobableSubstitutionModel |
| The EquiprobableSubstitutionModel substitution model for any kind of alphabet. Jukes-Cantor models are specific case of this model, applied to nucleotides or proteins. More... | |
| class | EventDrivenIntSymbolList |
| class | EvolutionSequenceSimulator |
| class | Exception |
| class | ExponentialDiscreteDistribution |
| class | ExponentialDiscreteRateDistribution |
| class | ExtendedFloat |
| class | ExtendedFloatArrayWrapper |
| class | ExtendedFloatCol |
| class | ExtendedFloatEigen |
| class | ExtendedFloatEigenBase |
| struct | ExtendedFloatEigenCore |
| class | ExtendedFloatNoAlias |
| class | ExtendedFloatRow |
| class | ExtendedFloatVectorwiseOp |
| class | ExtendedNewick |
| The so-called 'ExtendedNewick' parenthetic format for phylogenetic networks, where hybridization nodes are mandatory tagged and appear several times. More... | |
| class | F81 |
| The Felsenstein (1981) substitution model for nucleotides. More... | |
| class | F84 |
| The Felsenstein (1984) substitution model for nucleotides. More... | |
| class | Fasta |
| class | FileTools |
| class | FirstOrderDerivable |
| class | FirstOrderDerivableWrapper |
| class | FivePointsNumericalDerivative |
| class | FixedCodonFrequencySet |
| FrequencySet for codons, with no parameter. More... | |
| class | FixedFrequencySet |
| FrequencySet useful for homogeneous and stationary models. More... | |
| class | FixedNucleotideFrequencySet |
| FrequencySet useful for homogeneous and stationary models, nucleotide implementation. More... | |
| class | FixedProteinFrequencySet |
| FrequencySet useful for homogeneous and stationary models, protein implementation. More... | |
| class | Font |
| class | FontManager |
| class | ForwardHmmD2Likelihood_DF |
| class | ForwardHmmDLikelihood_DF |
| class | ForwardHmmLikelihood_DF |
| class | ForwardLikelihoodTree |
| class | FrequenciesFromFrequencySet |
| class | FrequenciesFromSimplex |
| class | FrequencySetInterface |
| Parametrize a set of state frequencies. More... | |
| class | FromMixtureSubstitutionModel |
| Model taken from a SubModel of a Mixture of SubstitutionModels. More... | |
| class | FromModelFrequencySet |
| FrequencySet defined from the equilibrium distribution of a given model. More... | |
| class | FullCodonFrequencySet |
| A generic FrequencySet for Full Codon alphabets. More... | |
| class | FullFrequencySet |
| A generic FrequencySet allowing for the estimation of all frequencies. More... | |
| class | FullHmmTransitionMatrix |
| class | FullNucleotideFrequencySet |
| Nucleotide FrequencySet using three independent parameters (theta, theta1, theta2) to modelize the four frequencies: More... | |
| class | FullPerAACodonFrequencySet |
| FrequencySet integrating ProteinFrequencySet inside CodonFrequencySet. In this case, FrequencieSet defined inside each amino acid is parametrized as a FullFrequencySet. Hence there are 61-20=41 parameters in addition of the parameters of the ProteinFrequencySet. More... | |
| class | FullProteinFrequencySet |
| Protein FrequencySet using 19 independent parameters to model the 20 frequencies. More... | |
| class | FunctionInterface |
| class | FunctionOperator |
| class | FunctionStopCondition |
| class | FunctionTools |
| class | FunctionWrapper |
| class | G2001 |
| Galtier's 2001 covarion model. More... | |
| class | GammaDiscreteDistribution |
| class | GammaDiscreteRateDistribution |
| class | GaussianDiscreteDistribution |
| class | GaussianDiscreteRateDistribution |
| class | gBGC |
| gBGC model. More... | |
| class | GCFrequencySet |
| Nucleotide FrequencySet using only one parameter, the GC content (denoted as 'GC.theta') More... | |
| class | GCPositionSubstitutionRegister |
| Distinguishes AT->GC vs GC->AT on given codon position (0, 1, or 2) More... | |
| class | GCSubstitutionRegister |
| Distinguishes AT<->GC from GC<->AT. More... | |
| class | GCSynonymousSubstitutionRegister |
| Distinguishes AT->GC vs GC->AT inside synonymous substitutions on third codon position. More... | |
| class | GenBank |
| class | GeneralSubstitutionRegister |
| Sets a Register based on a matrix of integers. If M is the matrix, M[i,j] is the number of the substitution type from i to j, or 0 if there is no substitution type from i to j. More... | |
| class | GeneticCode |
| class | GivenDataSubstitutionProcessSequenceSimulator |
| Sequences simulation under a unique substitution process, but with site specific posterior probabilities. More... | |
| class | GivenDataSubstitutionProcessSiteSimulator |
| Site simulation under a unique substitution process, given data. More... | |
| class | GlobalClockTreeLikelihoodFunctionWrapper |
| class | GlobalGraph |
| class | GoldenSectionSearch |
| class | GranthamAAChemicalDistance |
| class | GranthamAAPolarityIndex |
| class | GranthamAAVolumeIndex |
| class | Graph |
| class | GraphicDevice |
| class | GraphObserver |
| class | GTR |
| The General Time-Reversible substitution model for nucleotides. More... | |
| class | GY94 |
| The Goldman and Yang (1994) substitution model for codons. More... | |
| class | HierarchicalClustering |
| Hierarchical clustering. More... | |
| class | HKY85 |
| The Hasegawa M, Kishino H and Yano T (1985) substitution model for nucleotides. More... | |
| class | HmmBadStateException |
| class | HmmEmissionProbabilities |
| class | HmmEmissionProbabilities_Eigen |
| Interface for computing emission probabilities in a Hidden Markov Model. More... | |
| class | HmmLikelihood |
| class | HmmLikelihood_DF |
| A simple implementation of hidden Markov models recursion, in DataFlow implementation. More... | |
| class | HmmPhyloAlphabet |
| Hidden states alphabet. More... | |
| class | HmmPhyloEmissionProbabilities |
| Emission probabilities in the context of DF phylolikeihoods. More... | |
| class | HmmProcessAlphabet |
| Hidden states alphabet. More... | |
| class | HmmProcessPhyloLikelihood |
| Likelihood framework based on a hmm of simple likelihoods. More... | |
| class | HmmSequenceEvolution |
| Sequence evolution framework based on a hmm. More... | |
| class | HmmStateAlphabet |
| class | HmmTransitionMatrix |
| class | HmmUnvalidAlphabetException |
| class | HomogeneousTreeLikelihood |
| Specialization of the TreeLikelihood interface for the Homogeneous case. More... | |
| class | IAlignment |
| class | Identity |
| r = id(f) More... | |
| class | IDiscreteDistribution |
| class | IDistanceMatrix |
| class | IFrequencySet |
| General interface for distance matrix readers. More... | |
| class | IMultiPhyloDAG |
| General interface for multiple trees readers. More... | |
| class | IMultiPhyloTree |
| class | IMultiTree |
| General interface for multiple trees readers. More... | |
| class | IndexOutOfBoundsException |
| class | InfinityDerivableFirstOrderWrapper |
| class | InfinityDerivableSecondOrderWrapper |
| class | InfinityFunctionWrapper |
| class | InMixedSubstitutionModel |
| SubModel taken from a MixedTransitionModel, kept in the context of the MixedTransitionModel (see FromMixtureSubstitutionModel for an out of context subModel). So "rate" and "scale" are set for the MixedTransitionModel. More... | |
| class | InOrderTreeIterator |
| class | IntegerAlphabet |
| class | IntervalConstraint |
| class | IntervalTransformedParameter |
| class | IntSymbolList |
| class | IntSymbolListInterface |
| class | InvariantMixedDiscreteDistribution |
| class | InvertebrateMitochondrialGeneticCode |
| class | IODAG |
| General interface for DAG I/O. More... | |
| class | IoDiscreteDistribution |
| class | IoDiscreteDistributionFactory |
| class | IODistanceMatrix |
| class | IODistanceMatrixFactory |
| class | IOException |
| class | IOFormat |
| class | IoFrequencySet |
| General interface for model I/O. More... | |
| class | IOFrequencySetFactory |
| Utilitary class for creating frequencies set readers and writers. More... | |
| class | IOPairedSiteLikelihoods |
| Base class for I/O on paired-site likelihoods. More... | |
| class | IOParametrizable |
| class | IOPhymlPairedSiteLikelihoods |
| This class provides input for the Phyml paired-site likelihoods format. More... | |
| class | IOProbabilisticSequence |
| class | IOSequence |
| class | IoSequenceFactory |
| class | IOSequenceStream |
| class | IoSubstitutionModel |
| General interface for model I/O. More... | |
| class | IOSubstitutionModelFactory |
| Utilitary class for creating substitution model readers and writers. More... | |
| class | IOTree |
| General interface for tree I/O. More... | |
| class | IOTreeFactory |
| Utilitary class for creating tree readers and writers. More... | |
| class | IOTreepuzzlePairedSiteLikelihoods |
| This class provides I/O for the Tree-Puzzle/RAxML (phylip-like) paired-site likelihoods format. More... | |
| class | IPhyloDAG |
| General interface for DAG readers. More... | |
| class | IPhyloTree |
| General interface for tree readers. More... | |
| class | IProbabilisticAlignment |
| class | IProbabilisticSequence |
| class | ISequence |
| class | ISubstitutionModel |
| General interface for model readers. More... | |
| class | ITree |
| General interface for tree readers. More... | |
| class | JCnuc |
| The Jukes-Cantor substitution model for nucleotides. More... | |
| class | JCprot |
| The Jukes-Cantor substitution model for proteins. More... | |
| class | JTT92 |
| The Jones, Taylor and Thornton substitution model for proteins. More... | |
| class | K80 |
| The Kimura 2-rates substitution model for nucleotides. More... | |
| class | KCM |
| The general multiple substitution model for codons, from Zaheri & al, 2014. More... | |
| class | KD_AAHydropathyIndex |
| class | KeyvalException |
| class | KeyvalTools |
| class | KleinAANetChargeIndex |
| class | KrKcSubstitutionRegister |
| Count conservative and radical amino-acid substitutions. More... | |
| class | KroneckerCodonDistanceFrequenciesSubstitutionModel |
| Class for non-synonymous substitution models on codons with parameterized equilibrium frequencies and nucleotidic models, with allowed multiple substitutions. More... | |
| class | KroneckerCodonDistanceSubstitutionModel |
| Class for non-synonymous substitution models on codons with parameterized nucleotidic models, with allowed multiple substitutions. More... | |
| class | KroneckerWordSubstitutionModel |
| Basal class for words of substitution models with multiple substitutions. More... | |
| class | L95 |
| The no-strand bias substitution model for nucleotides, from Lobry 1995. The point of this model is that the substitution rate from a nucleotide N towards another M is the same as the rate from the complement of N towards the complement of M. Note that this model is not reversible. More... | |
| class | LabelCollapsedNodesTreeDrawingListener |
| A TreeDrawingListener implementation that label the collapsed nodes. More... | |
| class | LabelInnerNodesTreeDrawingListener |
| A TreeDrawingListener implementation that write the names of inner nodes. More... | |
| class | LabelSubstitutionCount |
| Labelling substitution count. More... | |
| class | LaplaceSubstitutionCount |
| Laplace estimate of the substitution count. More... | |
| class | LeafNamesTreeDrawingListener |
| A TreeDrawingListener implementation that write leaf names. More... | |
| class | LegacyAbstractMapping |
| Partial implementation of the mapping interface. More... | |
| class | LegacyAbstractRewardMapping |
| Partial implementation of the reward mapping interface. More... | |
| class | LegacyAbstractSubstitutionMapping |
| Partial implementation of the substitution mapping interface. More... | |
| class | LegacyAncestralStateReconstruction |
| Interface for ancestral states reconstruction methods. More... | |
| class | LegacyMappingInterface |
| General interface for storing mapping data. More... | |
| class | LegacyMarginalAncestralStateReconstruction |
| Likelihood ancestral states reconstruction: marginal method. More... | |
| class | LegacyOptimizationTools |
| Optimization methods for phylogenetic inference. More... | |
| class | LegacyPhylogeneticsApplicationTools |
| This class provides some common tools for applications. More... | |
| class | LegacyProbabilisticRewardMapping |
| Legacy data storage class for probabilistic rewards mappings. More... | |
| class | LegacyProbabilisticSubstitutionMapping |
| Legacy data storage class for probabilistic substitution mappings. More... | |
| class | LegacyRewardMappingInterface |
| Legacy interface for storing reward mapping data. More... | |
| class | LegacyRewardMappingTools |
| Provide methods to compute reward mappings. More... | |
| class | LegacySubstitutionMappingInterface |
| Legacy interface for storing mapping data. More... | |
| class | LegacySubstitutionMappingTools |
| Provide methods to compute substitution mappings. More... | |
| class | LetterAlphabet |
| class | LexicalAlphabet |
| class | LG08 |
| The Le and Gascuel substitution model for proteins. More... | |
| class | LG10_EX_EHO |
| The Le and Gascuel (2010) EX_EHO substitution model for proteins. More... | |
| class | LGL08_CAT |
| The Le et al (2008) CAT substitution model for proteins. More... | |
| class | LikelihoodCalculation |
| class | LikelihoodCalculationOnABranch |
| class | LikelihoodCalculationSingleProcess |
| class | LinearMatrix |
| class | LLG08_EHO |
| The Le et al (2008) EH0 substitution model for proteins. More... | |
| class | LLG08_EX2 |
| The Le et al (2008) EX2 substitution model for proteins. More... | |
| class | LLG08_EX3 |
| The Le et al (2008) EX3 substitution model for proteins. More... | |
| class | LLG08_UL2 |
| The Le et al (2008) UL2 substitution model for proteins. More... | |
| class | LLG08_UL3 |
| The Le et al (2008) UL3 substitution model for proteins. More... | |
| class | LogSumExp |
| class | LogsumHmmLikelihood |
| class | LowMemoryRescaledHmmLikelihood |
| class | LUDecomposition |
| class | MappedNamedContainer |
| class | MappingInterface |
| General interface for storing mapping data. More... | |
| class | MapTools |
| class | MarginalAncestralReconstruction |
| Likelihood ancestral states reconstruction: marginal method. More... | |
| class | MarkovModulatedFrequencySet |
| FrequencySet to be used with a Markov-modulated substitution model. More... | |
| class | MarkovModulatedStateMap |
| This class implements a state map for Markov modulated models. More... | |
| class | MarkovModulatedSubstitutionModel |
| Partial implementation of the Markov-modulated class of substitution models. More... | |
| class | Mase |
| class | MaseHeader |
| class | MaseTools |
| class | MathOperator |
| class | Matrix |
| struct | MatrixDimension |
| Basic matrix dimension type. More... | |
| class | MatrixProduct |
| class | MatrixTools |
| class | MetaOptimizer |
| class | MetaOptimizerInfos |
| class | MG94 |
| The Muse and Gaut (1994) substitution model for codons. More... | |
| class | MixedSubstitutionModelSet |
| Substitution models manager for Mixed Substitution Models. This class inherits from SubstitutionModelSet. More... | |
| class | MixedTransitionModelInterface |
| Interface for Transition models, defined as a mixture of "simple" transition models. More... | |
| class | MixtureOfASubstitutionModel |
| class | MixtureOfATransitionModel |
| Transition models defined as a mixture of nested substitution models. More... | |
| class | MixtureOfDiscreteDistributions |
| class | MixtureOfSubstitutionModels |
| class | MixtureOfTransitionModels |
| Transition models defined as a mixture of several substitution models. More... | |
| class | MixtureProcessPhyloLikelihood |
| Likelihood framework based on a mixture of simple likelihoods. More... | |
| class | MixtureSequenceEvolution |
| Sequence evolution framework based on a mixture of substitution processes. More... | |
| class | MiyataAAChemicalDistance |
| struct | ModelBranch |
| class | ModelList |
| A list of models, for building a WordSubstitutionModel. More... | |
| class | ModelPath |
| Organization of submodels in mixed substitution models in a path. See class ModelScenario for a thorough description. More... | |
| class | ModelScenario |
| Organization of submodels in mixed substitution models as paths. More... | |
| class | MoldMitochondrialGeneticCode |
| class | MolscriptColorSet |
| class | MultinomialFromTransitionModel |
| From a model, compute the likelihood of counts given an ancestral state. More... | |
| class | MultipleDiscreteDistribution |
| class | MultiProcessSequenceEvolution |
| Partial implementation of multiple processes of sequences. More... | |
| class | MultiProcessSequencePhyloLikelihood |
| Partial implementation of the Likelihood interface for multiple processes. More... | |
| class | MultiRange |
| class | MutationPath |
| This class is used by MutationProcess to store detailed results of simulations. More... | |
| class | MutationProcess |
| Interface for simulations. More... | |
| class | MvaFrequencySet |
| A frequencies set used to estimate frequencies at the root with the COaLA model. Frequencies at the root are optimized in the same way than the equlibrium frequencies on branches. Hyperparameters are used, which represent positions along the principal axes obtained from a preliminary Correspondence Analysis. From the optimized positions, the 20 frequencies are calculated. More... | |
| class | NaiveSubstitutionCount |
| Naive substitution count. More... | |
| class | NamedContainerInterface |
| class | NaNListener |
| A listener which capture NaN function values and throw an exception in case this happens. More... | |
| class | NegativeOperator |
| class | NeighborIteratorClass |
| class | NeighborJoining |
| The neighbor joining distance method. More... | |
| class | NestedStringTokenizer |
| class | Newick |
| The so-called 'newick' parenthetic format. More... | |
| class | NewtonBacktrackOneDimension |
| class | NewtonOneDimension |
| class | NexusIOSequence |
| class | NexusIOTree |
| a simple parser for reading trees from a Nexus file. More... | |
| class | NexusTools |
| class | Nhx |
| The so-called 'Nhx - New Hampshire eXtended' parenthetic format. More... | |
| class | NNIHomogeneousTreeLikelihood |
| This class adds support for NNI topology estimation to the DRHomogeneousTreeLikelihood class. More... | |
| class | NNISearchable |
| Interface for Nearest Neighbor Interchanges algorithms. More... | |
| class | NNITopologyListener |
| Listener used internally by the optimizeTreeNNI method. More... | |
| class | NNITopologyListener2 |
| Listener used internally by the optimizeTreeNNI2 method. More... | |
| class | NNITopologySearch |
| NNI topology search method. More... | |
| class | Node |
| The phylogenetic node class. More... | |
| class | Node_DF |
| Base dataflow Node class. More... | |
| class | NodeEvent |
| class | NodeException |
| General exception thrown when something is wrong with a particular node. More... | |
| class | NodeNotFoundException |
| Exception thrown when something is wrong with a particular node. More... | |
| class | NodePException |
| General exception thrown when something is wrong with a particular node. More... | |
| class | NodesIdTreeDrawingListener |
| A TreeDrawingListener implementation that writes nodes id. More... | |
| class | NodesIteratorClass |
| class | NodesIteratorClass< Graph::ALLGRAPHITER, is_const > |
| class | NodesIteratorClass< Graph::INCOMINGNEIGHBORITER, is_const > |
| class | NodesIteratorClass< Graph::OUTGOINGNEIGHBORITER, is_const > |
| class | NodeTemplate |
| The NodeTemplate class. More... | |
| struct | NoDimension |
| Empty type representing no dimensions. More... | |
| class | NoGapTemplateSiteContainerIterator |
| class | NonHomogeneousSequenceSimulator |
| (Legacy) Site and sequences simulation under non-homogeneous models. More... | |
| class | NonHomogeneousSubstitutionProcess |
| Substitution process manager for non-homogeneous / non-reversible models of evolution. More... | |
| class | NonHomogeneousTreeLikelihood |
| Specialization of the TreeLikelihood interface for the branch non-homogeneous and non-stationary models. More... | |
| class | NoTableColumnNamesException |
| class | NoTableRowNamesException |
| class | NotImplementedException |
| class | NucleicAcidsReplication |
| class | NucleicAlphabet |
| class | NucleicAlphabetState |
| class | NucleotideFrequencySetInterface |
| Parametrize a set of state frequencies for nucleotides. More... | |
| class | NucleotideReversibleSubstitutionModelInterface |
| Specialisation interface for rversible nucleotide substitution model. More... | |
| class | NucleotideSubstitutionModelInterface |
| Specialisation interface for nucleotide substitution model. More... | |
| class | NullOutputStream |
| class | NullPointerException |
| class | Number |
| class | NumberFormatException |
| class | NumCalcApplicationTools |
| class | NumConstants |
| struct | NumericalDependencyTransform |
| Template struct used to describe a dependency transformation before compute(). More... | |
| struct | NumericalDependencyTransform< Transposed< T > > |
| Implementation for a dependency transposition. More... | |
| struct | NumericalDerivativeConfiguration |
| Configuration for a numerical derivation: what delta to use, and type of derivation. More... | |
| class | NumericAlphabet |
| class | NumericConstant |
| r = constant_value. More... | |
| class | NumericMutable |
| r = variable_value. More... | |
| class | NumTools |
| class | OAlignment |
| class | ODiscreteDistribution |
| class | ODistanceMatrix |
| class | OFrequencySet |
| General interface for distance matrix writers. More... | |
| class | OMultiPhyloDAG |
| General interface for tree writers. More... | |
| class | OMultiPhyloTree |
| General interface for tree writers. More... | |
| class | OMultiTree |
| General interface for tree writers. More... | |
| class | OnABranchPhyloLikelihood |
| Wraps a dataflow graph as a function: resultNode = f(variableNodes). More... | |
| class | OneChangeRegisterTransitionModel |
| From a model, compute transition probabilities given there is at least a change of a category (ie a non null number in a register) in the branch. More... | |
| class | OneChangeTransitionModel |
| From a model, compute transition probabilities given there is at least a change in the branch. More... | |
| class | OneDimensionOptimizationTools |
| class | OneJumpSubstitutionCount |
| Computes the probability that at least one jump occurred on a branch, given the initial and final state. More... | |
| class | OneProcessSequenceEvolution |
| Evolution of a sequence performed by a unique SubstitutionProcess all along the sequence. More... | |
| class | OneProcessSequencePhyloLikelihood |
| The OneProcessSequencePhyloLikelihood class: phylogenetic likelihood computation with a single process. More... | |
| class | OneProcessSequenceSubstitutionMapping |
| The OneProcessSequenceSubstitutionMapping class: substitution mapping linked with a OneProcessSequencePhyloLikelihood. More... | |
| class | OParametrizable |
| class | Operator |
| class | OPhyloDAG |
| General interface for DAG writers. More... | |
| class | OPhyloTree |
| General interface for tree writers. More... | |
| class | OProbabilisticAlignment |
| class | OProbabilisticSequence |
| class | OptimizationEvent |
| class | OptimizationListener |
| class | OptimizationStopCondition |
| class | OptimizationTools |
| Optimization methods for phylogenetic inference. More... | |
| class | OptimizerInterface |
| class | OrderedSimplex |
| class | OSequence |
| class | OSubstitutionModel |
| General interface for distance matrix writers. More... | |
| class | OTree |
| General interface for tree writers. More... | |
| class | OutOfRangeException |
| class | OutputStream |
| class | PairedSiteLikelihoods |
| A container for paired-site likelihoods (likelihoods over the same sites for different models, especially topologies). An instance of this class is, roughly, a list of models, each of them having a name (stored in the modelNames attribute) and a set of site likelihoods (stored in the logLikelihoods attribute). More... | |
| class | Parameter |
| class | ParameterAliasable |
| class | ParameterAliasableAdapter |
| class | ParameterEvent |
| class | ParameterException |
| class | ParameterGrid |
| class | ParameterList |
| class | ParameterListener |
| class | ParameterNotFoundException |
| class | ParametersStopCondition |
| class | Parametrizable |
| class | ParametrizableAdapter |
| class | ParametrizableCollection |
| class | ParametrizablePhyloTree |
| PhyloTree with Parametrizable Phylo Branches. They SHARE their branch length parameters. More... | |
| class | PartitionProcessPhyloLikelihood |
| class | PartitionSequenceEvolution |
| Sequence evolution framework based on a mixture of substitution processes. More... | |
| class | Pasta |
| class | PatternTools |
| Utilitary methods to compute site patterns. More... | |
| class | PgfGraphicDevice |
| class | PGMA |
| Compute WPGMA and UPGMA trees from a distance matrix. More... | |
| struct | PGMAInfos |
| Inner data structure for WPGMA and UPGMA distance methods. More... | |
| class | PhredPhd |
| class | PhredPoly |
| class | Phylip |
| class | PhylipDistanceMatrixFormat |
| class | PhyloBranch |
| class | PhyloBranchException |
| General exception thrown when something is wrong with a particular branch. More... | |
| class | PhyloBranchMapping |
| class | PhyloBranchNotFoundException |
| Exception thrown when something is wrong with a particular branch. More... | |
| class | PhyloBranchParam |
| class | PhyloBranchPException |
| General exception thrown when something is wrong with a particular branch. More... | |
| class | PhyloBranchPropertyNotFoundException |
| General exception thrown if a property could not be found. More... | |
| class | PhyloBranchReward |
| class | PhyloDAG |
| class | PhylogeneticsApplicationTools |
| This class provides some common tools for applications. More... | |
| class | PhylogramDrawBranchEvent |
| class | PhylogramPlot |
| Phylogram representation of trees. More... | |
| class | PhyloLikelihoodContainer |
| The PhyloLikelihoodContainer, owns and assigns numbers to Phylolikelihoods. More... | |
| class | PhyloLikelihoodFormula |
| The PhyloLikelihoodFormula class, for phylogenetic likelihood on several independent data. More... | |
| class | PhyloLikelihoodInterface |
| The PhyloLikelihood interface, for phylogenetic likelihood. More... | |
| class | PhyloLikelihoodSetInterface |
| The PhyloLikelihoodSet interface, to manage a subset of PhyloLikelihoods from a given PhyloLikelihoodContainer. More... | |
| class | PhyloNode |
| class | PhyloNodeException |
| General exception thrown when something is wrong with a particular node. More... | |
| class | PhyloNodeNotFoundException |
| Exception thrown when something is wrong with a particular node. More... | |
| class | PhyloNodePException |
| General exception thrown when something is wrong with a particular node. More... | |
| class | PhyloNodePropertyNotFoundException |
| General exception thrown if a property could not be found. More... | |
| class | PhyloStatistics |
| Compute several quantities on a tree simulateously, optimizing the recursions on the tree. More... | |
| class | PhyloSubstitutionMapping |
| class | PhyloTree |
| class | PhyloTreeException |
| General exception thrown when something wrong happened in a tree. More... | |
| class | PhyloTreeTools |
| Generic utilitary methods dealing with trees. More... | |
| class | PlaceboTransformedParameter |
| class | Point2D |
| class | POMO |
| class | PositionedContainerInterface |
| class | PositionedNamedContainerInterface |
| class | PostOrderTreeIterator |
| class | PowellMultiDimensions |
| class | PreOrderTreeIterator |
| class | PrincipalComponentAnalysis |
| class | ProbabilisticRewardMapping |
| Data storage class for probabilistic rewards mappings. More... | |
| class | ProbabilisticSequence |
| class | ProbabilisticSequenceInterface |
| class | ProbabilisticSite |
| class | ProbabilisticSiteInterface |
| class | ProbabilisticSubstitutionMapping |
| Data storage class for probabilistic substitution mappings. More... | |
| class | ProbabilisticSymbolList |
| class | ProbabilisticSymbolListInterface |
| class | ProbabilitiesFromDiscreteDistribution |
| class | ProbabilitiesFromMixedModel |
| class | ProbabilityDAG |
| class | ProbabilityFromDiscreteDistribution |
| class | ProbabilityFromMixedModel |
| class | ProcessComputationEdge |
| class | ProcessComputationNode |
| Tree Organization of Computing Nodes. More... | |
| class | ProcessComputationTree |
| class | ProcessEdge |
| class | ProcessTree |
| struct | ProcPos |
| Likelihood framework based on a partition of a sequence in simple likelihoods. More... | |
| class | PropertyNotFoundException |
| General exception thrown if a property could not be found. More... | |
| class | ProteicAlphabet |
| class | ProteicAlphabetIndex1 |
| class | ProteicAlphabetIndex2 |
| class | ProteicAlphabetState |
| class | ProteinFrequencySetInterface |
| Parametrize a set of state frequencies for proteins. More... | |
| class | ProteinReversibleSubstitutionModelInterface |
| Specialized interface for protein reversible substitution model. More... | |
| class | ProteinSubstitutionModelInterface |
| Specialized interface for protein substitution model. More... | |
| class | PseudoNewtonOptimizer |
| This Optimizer implements Newton's algorithm for finding a minimum of a function. This is in fact a modified version of the algorithm, as suggested by Nicolas Galtier, for the purpose of optimizing phylogenetic likelihoods. More... | |
| class | RandomTools |
| class | Range |
| class | RangeCollection |
| class | rangeComp_ |
| class | RangeSet |
| class | RASiteSimulationResult |
| Data structure to store the result of a DetailedSiteSimulator. More... | |
| class | RASTools |
| Tools to deal with Rates Across Sites (RAS) models. More... | |
| class | RateAcrossSitesSubstitutionProcess |
| class | RColorSet |
| class | RE08 |
| The Rivas-Eddy substitution model with gap characters. More... | |
| class | RE08Codon |
| This is a wrapper class of RE08 for codon substitution models. More... | |
| class | RE08Nucleotide |
| This is a wrapper class of RE08 for nucleotide substitution models. More... | |
| class | RE08Protein |
| This is a wrapper class of RE08 for protein substitution models. More... | |
| struct | ReductionOf |
| class | RegisterRatesSubstitutionModel |
| From a model, substitution rates are set into categories following a given register. Each substitution of a category is then multiplied by a rate parameter specific to this category. More... | |
| class | RELAX |
| The RELAX (2014) branch-site model for codons. More... | |
| class | ReparametrizationDerivableFirstOrderWrapper |
| class | ReparametrizationDerivableSecondOrderWrapper |
| class | ReparametrizationFunctionWrapper |
| class | RescaledHmmLikelihood |
| class | ReverseTransliteratorInterface |
| class | ReversibleSubstitutionModelInterface |
| Interface for reversible substitution models. More... | |
| class | Reward |
| The Reward interface. More... | |
| class | RewardMappingInterface |
| General interface for storing reward mapping data. More... | |
| class | RewardMappingTools |
| Provide methods to compute reward mappings. More... | |
| class | RGBColor |
| class | RHomogeneousMixedTreeLikelihood |
| class | RHomogeneousTreeLikelihood |
| This class implement the 'traditional' way of computing likelihood for a tree. More... | |
| class | RN95 |
| The model described by Rhetsky & Nei, where the only hypothesis is that the transversion rates are only dependent of the target nucleotide. This model is not reversible. More... | |
| class | RN95s |
| Intersection of models RN95 and L95. More... | |
| class | RNA |
| class | RNonHomogeneousMixedTreeLikelihood |
| class | RNonHomogeneousTreeLikelihood |
| This class implement the 'traditional' way of computing likelihood for a tree, allowing for non-homogeneous models of substitutions. More... | |
| class | RNY |
| class | RowMatrix |
| struct | RowVectorDimension |
| class | RTransformedParameter |
| class | ScalarProduct |
| class | SecondOrderDerivable |
| class | SecondOrderDerivableWrapper |
| class | SelectedSubstitutionRegister |
| Class inheriting from GeneralSubstitutionRegister, this one uses a special constructor which allows it to build a substitution matrix from string input specifying a desired substitutions. More... | |
| class | SelfMutationProcess |
| This class is mainly for testing purpose. It allow "self" mutation of the kind i->i;. More... | |
| class | SENCA |
| Class for non-synonymous and synonymous substitution models on codons with parameterized equilibrium frequencies and nucleotidic basic models. More... | |
| class | Sequence |
| class | Sequence_DF |
| Data flow node representing a Sequence as a Value<Eigen::MatrixXd> with a name. More... | |
| class | SequenceAnnotation |
| class | SequenceApplicationTools |
| class | SequenceContainerTools |
| class | SequencedValuesContainer |
| class | SequenceEvolution |
| This interface describes the evolution process of a sequence. More... | |
| class | SequenceException |
| class | SequenceFileIndex |
| class | SequenceInterface |
| class | SequenceMask |
| class | SequenceNotAlignedException |
| class | SequenceNotFoundException |
| class | SequencePhyloLikelihoodInterface |
| PhyloLikelihoods based on Sequence Evolution class, ie for which there is a (set of) processes able to build a sequence. More... | |
| class | SequencePositionIterator |
| class | SequenceQuality |
| class | SequenceSimulationTools |
| Tools for sites and sequences simulation. More... | |
| class | SequenceSimulatorInterface |
| The SequenceSimulator interface. SequenceSimulator classes can simulate whole datasets. More... | |
| class | SequenceTools |
| class | SequenceWalker |
| class | SequenceWithAnnotation |
| class | SequenceWithAnnotationTools |
| class | SequenceWithGapException |
| class | SequenceWithQuality |
| class | SequenceWithQualityTools |
| class | ShiftDelta |
| class | ShiftParameter |
| shift param value = n * delta + x. More... | |
| class | SimData |
| class | SimpleCommentable |
| class | SimpleDiscreteDistribution |
| class | SimpleIndexDistance |
| class | SimpleMultiDimensions |
| class | SimpleMutationProcess |
| Generally used mutation process model. More... | |
| class | SimpleNewtonMultiDimensions |
| class | SimpleScore |
| class | SimpleSequencePositionIterator |
| class | SimpleSubstitutionProcess |
| Space and time homogeneous substitution process, without mixture. More... | |
| class | SimpleSubstitutionProcessSequenceSimulator |
| Sequences simulation under a unique substitution process. More... | |
| class | SimpleSubstitutionProcessSiteSimulator |
| Site simulation under a unique substitution process. More... | |
| class | SimpleTemplateSiteContainerIterator |
| class | Simplex |
| class | SimProcessEdge |
| class | SimProcessNode |
| class | SingleDataPhyloLikelihoodInterface |
| The SingleDataPhyloLikelihood interface, for phylogenetic likelihood. More... | |
| class | SingleProcessPhyloLikelihood |
| Wraps a dataflow graph as a function: resultNode = f(variableNodes). More... | |
| class | SingleProcessSubstitutionMapping |
| The SingleProcessSubstitutionMapping class: substitution mapping linked with a SingleProcessPhyloLikelihood. More... | |
| class | Site |
| class | SiteContainerTools |
| class | SiteException |
| class | SiteInterface |
| class | SiteNotFoundException |
| class | SitePartition |
| This is the interface for classes describing a site partition, each partition being intended to have its own substitution model. More... | |
| class | SitePartitionHomogeneousTreeLikelihood |
| Specialization of the TreeLikelihood interface for partition models, homogeneous case. More... | |
| class | SitePatterns |
| Data structure for site patterns. More... | |
| class | SiteSimulationResult |
| Data structure to store the result of a DetailedSiteSimulator. More... | |
| class | SiteSimulatorInterface |
| The SiteSimulator interface. SiteSimulator classes can simulate single sites. More... | |
| class | SiteTools |
| class | SiteWithGapException |
| class | SSR |
| The Strand Symmetric Reversible substitution model for nucleotides. More... | |
| class | StandardGeneticCode |
| class | StateChangedEvent |
| class | StateListener |
| class | StateMapInterface |
| Map the states of a given alphabet which have a model state. More... | |
| class | StatTest |
| class | StatTools |
| class | StdErr |
| class | StdOut |
| class | StdStr |
| class | StlOutputStream |
| class | StlOutputStreamWrapper |
| class | StochasticMapping |
| class | Stockholm |
| class | StopCodonException |
| class | StringSequenceTools |
| class | StringTokenizer |
| class | SubstitutionCountInterface |
| The SubstitutionsCount interface. More... | |
| class | SubstitutionDistance |
| Interface allowing for using distances between states in substitution counts. More... | |
| class | SubstitutionMapping |
| General interface for storing mapping data. More... | |
| class | SubstitutionMappingTools |
| Provide methods to compute substitution mappings. More... | |
| class | SubstitutionModelInterface |
| Interface for all substitution models. More... | |
| class | SubstitutionModelSet |
| Substitution models manager for non-homogeneous / non-reversible models of evolution. More... | |
| class | SubstitutionModelSetTools |
| Tools for automatically creating SubstitutionModelSet objects. More... | |
| class | SubstitutionProcessCollection |
| Collection of Substitution Process, which owns all the necessary objects: Substitution models, frequencies sets, rate distributions and trees. More... | |
| class | SubstitutionProcessCollectionMember |
| class | SubstitutionProcessInterface |
| This interface describes the substitution process along the tree and sites of the alignment. More... | |
| class | SubstitutionProcessSequenceSimulator |
| Sequences simulation under position specific substitution process. More... | |
| class | SubstitutionRegisterInterface |
| The SubstitutionRegister interface. More... | |
| class | SumOfLogarithms |
| class | SvgGraphicDevice |
| class | SwitchDeleter |
| class | SWSubstitutionRegister |
| Distinguishes substitutions given the link between the changed nucleotides : S for strong (GC) and W for weak (AT). More... | |
| class | SymbolListTools |
| class | T92 |
| The Tamura (1992) substitution model for nucleotides. More... | |
| class | Table |
| class | TableColumnNameNotFoundException |
| class | TableColumnNamesException |
| class | TableNameNotFoundException |
| class | TableRowNameNotFoundException |
| class | TableRowNamesException |
| class | TemplateAlignedSequenceContainer |
| class | TemplateAlignmentDataInterface |
| class | TemplateCoreSymbolListInterface |
| class | TemplateEventDrivenCoreSymbolListInterface |
| class | TemplateISequenceStream |
| class | TemplateOSequenceStream |
| class | TemplateSequenceContainerInterface |
| class | TemplateSequenceDataInterface |
| class | TemplateSequenceIteratorInterface |
| class | TemplateSiteContainerInterface |
| class | TemplateSiteIteratorInterface |
| class | TemplateStreamSequenceIterator |
| class | TemplateVectorSequenceContainer |
| class | TemplateVectorSiteContainer |
| class | TestFunction |
| class | ThreePointsNumericalDerivative |
| class | TN93 |
| The Tamura and Nei (1993) substitution model for nucleotides. More... | |
| class | TopologyChangeEvent |
| Class for notifying new toplogy change events. More... | |
| class | TopologyListener |
| Implement this interface to be notified when the topology of a tree has changed during topology search. More... | |
| class | TopologySearch |
| Interface for topology search methods. More... | |
| class | TotalSubstitutionRegister |
| Count all substitutions. More... | |
| class | TransformedParameter |
| class | TransitionFromTransitionModel |
| From a transition model, compute the transition function probabilities. More... | |
| class | TransitionFunctionFromModel |
| class | TransitionMatrixFromModel |
| class | TransitionMatrixFromTransitionMatrix |
| class | TransitionModelInterface |
| Interface for all transition models. More... | |
| class | TransliteratorInterface |
| struct | Transposed |
| The T dependency should be transposed before computation. More... | |
| class | Tree |
| Interface for phylogenetic tree objects. More... | |
| class | TreeDrawing |
| Basal interface for tree drawing classes. More... | |
| class | TreeDrawingDisplayControler |
| Easy tune of tree drawings display. More... | |
| class | TreeDrawingListener |
| Interface allowing to capture drawing events. More... | |
| class | TreeDrawingListenerAdapter |
| An empty implementation of the TreeDrawingListener interface. More... | |
| class | TreeDrawingNodeInfo |
| class | TreeDrawingSettings |
| A set of options to tune the display of a TreeDrawing object. More... | |
| class | TreeException |
| General exception thrown when something wrong happened in a tree. More... | |
| class | TreeGraph |
| class | TreeGraphImpl |
| class | TreeIterator |
| class | TreeLikelihoodData |
| TreeLikelihood data structure. More... | |
| class | TreeLikelihoodInterface |
| The TreeLikelihood interface. More... | |
| class | TreeLikelihoodNodeData |
| TreeLikelihood partial data structure. More... | |
| class | TreeLikelihoodTools |
| Utilitary methods that work with TreeLikelihood objects. More... | |
| class | TreeParsimonyDataInterface |
| TreeParsimonyScore data structure. More... | |
| class | TreeParsimonyNodeDataInterface |
| TreeParsimonyScore node data structure. More... | |
| class | TreeParsimonyScoreInterface |
| Compute a parsimony score. More... | |
| class | TreeTemplate |
| The phylogenetic tree class. More... | |
| class | TreeTemplateTools |
| Utilitary methods working with TreeTemplate and Node objects. More... | |
| class | TreeTools |
| Generic utilitary methods dealing with trees. More... | |
| class | TripletSubstitutionModel |
| Class for neutral substitution models on triplets, which correspond to codons that do not have any significance (whether they are STOP or functional). More... | |
| class | TrivialSitePartition |
| Trivial site partition: all sites belong to the same, unique partition. More... | |
| class | TruncatedExponentialDiscreteDistribution |
| class | TS98 |
| Tuffley and Steel's 1998 covarion model. More... | |
| class | TsTvSubstitutionRegister |
| Distinguishes transitions from transversions. More... | |
| class | TwoParameterBinarySubstitutionModel |
| The Model on two states. More... | |
| class | TwoPointsNumericalDerivative |
| class | UniformDiscreteDistribution |
| class | UniformizationSubstitutionCount |
| Analytical (weighted) substitution count using the uniformization method. More... | |
| class | UnrootedPhyloTreeException |
| Exception thrown when a tree is expected to be rooted. More... | |
| class | UnrootedTreeException |
| Exception thrown when a tree is expected to be rooted. More... | |
| class | UnvalidFlagException |
| class | UserAlphabetIndex1 |
| class | UserAlphabetIndex2 |
| class | UserCodonFrequencySet |
| class | UserFrequencySet |
| FrequencySet to be read in a file. More specifically, a frequency set is read in a column of a given file, which column number is given in argument (default: 1). More... | |
| class | UserNucleotideFrequencySet |
| FrequencySet useful for homogeneous and stationary models, nucleotide implementation. More... | |
| class | UserProteinFrequencySet |
| FrequencySet from file. More... | |
| class | UserProteinSubstitutionModel |
| Build an empirical protein substitution model from a file. More... | |
| class | Value |
| Abstract Node storing a value of type T. More... | |
| class | ValueFromConfiguredParameter |
| struct | VectorDimension |
| class | VectorException |
| class | VectorMappedContainer |
| class | VectorOfSubstitutionRegisters |
| Sets a Register based on a vector of Registers. The categories are intersection of categories of those Registers. More... | |
| class | VectorPositionedContainer |
| class | VectorTools |
| class | VertebrateMitochondrialGeneticCode |
| class | WAG01 |
| The Whelan and Goldman substitution model for proteins. More... | |
| class | WeightedSubstitutionCount |
| Interface allowing for weighting of substitution counts according to state properties. More... | |
| class | WordAlphabet |
| class | WordFrequencySetInterface |
| Frequencies in words computed from the frequencies on letters. The parameters are the parameters of the Frequencies on letters. The WordFrequencySet owns the std::shared_ptr<FrequencySet> it is built on. Interface class. More... | |
| class | WordFromIndependentFrequencySet |
| the Frequencies in words are the product of Independent Frequencies in letters More... | |
| class | WordFromUniqueFrequencySet |
| class | WordSubstitutionModel |
| Basal class for words of substitution models. More... | |
| class | WrappedModelInterface |
| Wrapping model interface. More... | |
| class | WrappedSubstitutionModelInterface |
| class | WrappedTransitionModelInterface |
| class | XFigColorManager |
| class | XFigGraphicDevice |
| class | XFigLaTeXFontManager |
| class | XFigPostscriptFontManager |
| class | YeastMitochondrialGeneticCode |
| class | YN98 |
| The Yang and Nielsen (1998) substitution model for codons. More... | |
| class | YNGP_M |
| Abstract generic class for The Yang et al (2000) M substitution models for codons. al (2004). More... | |
| class | YNGP_M1 |
| The Yang et al (2000) M1 substitution model for codons, with the more realistic modification in Wong & al (2004). More... | |
| class | YNGP_M10 |
| The Yang et al (2000) M10 substitution model for codons. More... | |
| class | YNGP_M2 |
| The Yang et al (2000) M2 substitution model for codons, with the more realistic modification in Wong & al (2004). More... | |
| class | YNGP_M3 |
| The Yang et al (2000) M3 substitution model for codons. More... | |
| class | YNGP_M7 |
| The Yang et al (2000) M7 substitution model for codons. More... | |
| class | YNGP_M8 |
| The Yang et al (2000) M8 substitution model for codons. More... | |
| class | YNGP_M9 |
| The Yang et al (2000) M9 substitution model for codons. More... | |
| class | YpR |
| YpR model. More... | |
| class | YpR_Gen |
| General YpR model. More... | |
| class | YpR_Sym |
| symmetrical YpR model. More... | |
| class | ZeroDivisionException |
Enumerations | |
| enum class | DotOptions { None = 0 , DetailedNodeInfo = 1 << 0 , FollowUpwardLinks = 1 << 1 , ShowDependencyIndex = 1 << 2 } |
| enum class | NumericalProperty { Constant , ConstantZero , ConstantOne , ConstantIdentity } |
| Numerical properties for DF Node. More... | |
| enum class | NumericalDerivativeType { Disabled , ThreePoints , FivePoints } |
Functions | |
| std::ostream & | operator<< (std::ostream &out, const BppBoolean &s) |
| std::ostream & | operator<< (std::ostream &out, const BppString &s) |
| bool | operator== (const Matrix< Scalar > &m1, const Matrix< Scalar > &m2) |
| std::vector< T > | operator+ (const std::vector< T > &v1, const std::vector< T > &v2) |
| std::vector< T > | operator- (const std::vector< T > &v1, const std::vector< T > &v2) |
| std::vector< T > | operator* (const std::vector< T > &v1, const std::vector< T > &v2) |
| std::vector< T > | operator/ (const std::vector< T > &v1, const std::vector< T > &v2) |
| std::vector< T > | operator+ (const std::vector< T > &v1, const C &c) |
| std::vector< T > | operator+ (const C &c, const std::vector< T > &v1) |
| std::vector< T > | operator- (const std::vector< T > &v1, const C &c) |
| std::vector< T > | operator- (const C &c, const std::vector< T > &v1) |
| std::vector< T > | operator* (const std::vector< T > &v1, const C &c) |
| std::vector< T > | operator* (const C &c, const std::vector< T > &v1) |
| std::vector< T > | operator/ (const std::vector< T > &v1, const C &c) |
| std::vector< T > | operator/ (const C &c, const std::vector< T > &v1) |
| void | operator+= (std::vector< T > &v1, const std::vector< T > &v2) |
| void | operator-= (std::vector< T > &v1, const std::vector< T > &v2) |
| void | operator*= (std::vector< T > &v1, const std::vector< T > &v2) |
| void | operator/= (std::vector< T > &v1, const std::vector< T > &v2) |
| void | operator&= (std::vector< T > &v1, const C &c) |
| void | operator+= (std::vector< T > &v1, const C &c) |
| void | operator-= (std::vector< T > &v1, const C &c) |
| void | operator*= (std::vector< T > &v1, const C &c) |
| void | operator/= (std::vector< T > &v1, const C &c) |
| std::string | prettyTypeName (const std::type_info &type_info) |
| Debug: return a readable name for a C++ type descriptor (from typeid operator). More... | |
| bool | isTransitivelyDependentOn (const Node_DF &searchedDependency, const Node_DF &node) |
| Check if searchedDependency if a transitive dependency of node. More... | |
| NodeRef | recreateWithSubstitution (Context &c, const NodeRef &node, const std::unordered_map< const Node_DF *, NodeRef > &substitutions) |
| Recreate node by transitively replacing dependencies according to substitutions mapping. More... | |
| DotOptions | operator| (DotOptions a, DotOptions b) |
| bool | operator& (DotOptions a, DotOptions b) |
| static std::string | dotLabelEscape (const std::string &s) |
| static std::string | dotIdentifier (const Node_DF &node) |
| static void | writeDotNode (std::ostream &os, const Node_DF &node, DotOptions opt) |
| static void | writeDotEdge (std::ostream &os, const Node_DF &from, std::size_t depIndex, DotOptions opt) |
| static void | writeGraphStructure (std::ostream &os, const std::vector< const Node_DF * > &entryPoints, DotOptions opt) |
| void | writeGraphToDot (std::ostream &os, const std::vector< const Node_DF * > &nodes, DotOptions opt=DotOptions::None) |
| Write dataflow graph starting at nodes to output stream. More... | |
| void | writeGraphToDot (const std::string &filename, const std::vector< const Node_DF * > &nodes, DotOptions opt=DotOptions::None) |
| Write dataflow graph starting at nodes to file at filename, overwriting it. More... | |
| template<typename T > | |
| void | combineHash (std::size_t &seed, const T &t) |
| Combine hashable value to a hash, from Boost library. More... | |
| template<typename T > | |
| std::string | to_string_with_precision (const T a_value, const int n=6) |
| Output with given precision. More... | |
| template<typename T , typename U > | |
| std::shared_ptr< T > | convertRef (const std::shared_ptr< U > &from) |
| Convert a node ref with runtime type check. More... | |
| template<typename T > | |
| const T & | accessValueConstCast (const Node_DF &node) |
| template<typename T > | |
| std::shared_ptr< T > | cachedAs (Context &c, std::shared_ptr< T > &&newNode) |
| Helper: Same as Context::cached but with a shared_ptr<T> node. More... | |
| template<typename T > | |
| std::shared_ptr< T > | cachedAs (Context &c, std::shared_ptr< T > &newNode) |
| void | failureDeltaNotDerivable (const std::type_info &contextNodeType) |
| void | failureNumericalDerivationNotConfigured () |
| template<typename NodeT , typename DepT , typename B > | |
| ValueRef< NodeT > | generateNumericalDerivative (Context &c, const NumericalDerivativeConfiguration &config, NodeRef dep, const Dimension< DepT > &depDim, Dimension< NodeT > &nodeDim, B buildNodeWithDep) |
| Helper used to generate data flow expressions computing a numerical derivative. More... | |
| template<typename NodeT , typename B > | |
| ValueRef< NodeT > | generateNumericalDerivative (Context &c, const NumericalDerivativeConfiguration &config, NodeRef dep, const Dimension< NodeT > &nodeDim, B buildNodeWithDep) |
| template void | copyBppToEigen (const std::vector< ExtendedFloat > &bppVector, Eigen::RowVectorXd &eigenVector) |
| template void | copyBppToEigen (const std::vector< ExtendedFloat > &bppVector, Eigen::VectorXd &eigenVector) |
| template void | copyBppToEigen (const std::vector< ExtendedFloat > &bppVector, ExtendedFloatRowVectorXd &eigenVector) |
| template void | copyBppToEigen (const bpp::Vdouble &bppVector, Eigen::RowVectorXd &eigenVector) |
| template void | copyBppToEigen (const bpp::Vdouble &bppVector, Eigen::VectorXd &eigenVector) |
| template void | copyBppToEigen (const bpp::Vdouble &bppVector, ExtendedFloatRowVectorXd &eigenVector) |
| void | copyBppToEigen (const std::vector< ExtendedFloatVectorXd > &bppVector, ExtendedFloatMatrixXd &eigenVector) |
| void | copyBppToEigen (const std::vector< Eigen::VectorXd > &bppVector, Eigen::MatrixXd &eigenMatrix) |
| void | copyBppToEigen (const bpp::Matrix< double > &bppMatrix, ExtendedFloatMatrixXd &eigenMatrix) |
| void | copyBppToEigen (const bpp::Matrix< double > &bppMatrix, Eigen::MatrixXd &eigenMatrix) |
| template void | copyEigenToBpp (const ExtendedFloatMatrixXd &eigenMatrix, std::vector< std::vector< double > > &bppMatrix) |
| template void | copyEigenToBpp (const ExtendedFloatMatrixXd &eigenMatrix, std::vector< std::vector< bpp::ExtendedFloat > > &bppMatrix) |
| template void | copyEigenToBpp (const Eigen::MatrixXd &eigenMatrix, std::vector< std::vector< double > > &bppMatrix) |
| template void | copyEigenToBpp (const Eigen::MatrixXd &eigenMatrix, std::vector< std::vector< bpp::ExtendedFloat > > &bppMatrix) |
| void | copyEigenToBpp (const MatrixLik &eigenMatrix, bpp::Matrix< double > &bppMatrix) |
| template void | copyEigenToBpp (const RowLik &eigenVector, Vdouble &bppVector) |
| template void | copyEigenToBpp (const RowLik &eigenVector, std::vector< ExtendedFloat > &bppVector) |
| std::string | to_string (const NoDimension &) |
| std::string | to_string (const MatrixDimension &dim) |
| std::size_t | hash (const MatrixDimension &dim) |
| void | checkRecreateWithoutDependencies (const std::type_info &contextNodeType, const NodeRefVec &deps) |
| template<typename eVector , typename bVector , ForConvert< eVector, bVector > = true> | |
| void | copyBppToEigen (const bVector &bppVector, eVector &eigenVector) |
| template<typename eVector , EFVector< eVector > = true> | |
| void | copyBppToEigen (const std::vector< ExtendedFloat > &bppVector, eVector &eigenVector) |
| template<typename eVector , EFoutput< eVector > = true> | |
| void | copyBppToEigen (const Vdouble &bppVector, eVector &eigenVector) |
| template<typename eType , typename = typename std::enable_if<(std::is_same<eType, double>::value || std::is_same<eType, ExtendedFloat>::value)>::type> | |
| void | copyEigenToBpp (const ExtendedFloatMatrixXd &eigenMatrix, std::vector< std::vector< eType > > &bppMatrix) |
| template<typename eType , typename = typename std::enable_if<(std::is_same<eType, double>::value || std::is_same<eType, ExtendedFloat>::value)>::type> | |
| void | copyEigenToBpp (const Eigen::MatrixXd &eigenMatrix, std::vector< std::vector< eType > > &bppMatrix) |
| template<typename eVector , typename bVector , ForConvert2< bVector, eVector > = true> | |
| void | copyEigenToBpp (const bVector &eigenVector, eVector &bppVector) |
| std::size_t | hash (const NoDimension &) |
| bool | operator== (const NoDimension &, const NoDimension &) |
| bool | operator!= (const NoDimension &, const NoDimension &) |
| bool | operator== (const MatrixDimension &lhs, const MatrixDimension &rhs) |
| bool | operator!= (const MatrixDimension &lhs, const MatrixDimension &rhs) |
| template<typename Predicate > | |
| void | removeDependenciesIf (NodeRefVec &deps, Predicate p) |
| template<typename T > | |
| constexpr T | constexpr_power (T d, int n) |
| int | powi (int base, unsigned int exp) |
| std::ostream & | operator<< (std::ostream &os, const ExtendedFloat &ef) |
| std::string | to_string (const ExtendedFloat &ef) |
| double | log (const ExtendedFloat &ef) |
| ExtendedFloat | operator* (const double &lhs, const ExtendedFloat &rhs) |
| ExtendedFloat | operator- (const double &lhs, const ExtendedFloat &rhs) |
| ExtendedFloat | operator+ (const double &lhs, const ExtendedFloat &rhs) |
| ExtendedFloat | operator/ (const double &lhs, const ExtendedFloat &rhs) |
| ExtendedFloat | pow (const ExtendedFloat &ef, double exp) |
| ExtendedFloat | exp (const ExtendedFloat &ef) |
| ExtendedFloat | abs (const ExtendedFloat &ef) |
| double | convert (const bpp::ExtendedFloat &ef) |
| template<int R, int C> | |
| ExtendedFloatArray< R, C > | log (const ExtendedFloatArray< R, C > &arr) |
| template<int R, int C, template< int R2=R, int C2=C > class MatType> | |
| ExtendedFloatEigen< R, C, MatType > | exp (const ExtendedFloatEigen< R, C, MatType > &arr) |
| template<int R, int C> | |
| ExtendedFloatArray< R, C > | pow (const ExtendedFloatArray< R, C > &obj, double exp) |
| template<int R, int C> | |
| ExtendedFloatArray< R, C > | pow (const ExtendedFloatArray< R, C > &obj, int exp) |
| template<int R, int C, template< int R2=R, int C2=C > class MatType, typename T > | |
| std::enable_if< std::is_same< T, ExtendedFloat >::value||std::is_floating_point< T >::value||std::is_integral< T >::value, ExtendedFloatEigen< R, C, MatType > >::type | operator+ (const T &d, const ExtendedFloatEigen< R, C, MatType > rhs) |
| template<int R, int C, template< int R2=R, int C2=C > class MatType, typename T > | |
| std::enable_if< std::is_same< T, ExtendedFloat >::value||std::is_floating_point< T >::value||std::is_integral< T >::value, ExtendedFloatEigen< R, C, MatType > >::type | operator- (const T &d, const ExtendedFloatEigen< R, C, MatType > rhs) |
| template<int R, int C, template< int R2=R, int C2=C > class MatType, typename T > | |
| std::enable_if< std::is_same< T, ExtendedFloat >::value||std::is_floating_point< T >::value||std::is_integral< T >::value, ExtendedFloatEigen< R, C, MatType > >::type | operator* (const T &fact, const ExtendedFloatEigen< R, C, MatType > mat) |
| template<typename Derived , typename EFType > | |
| EFType | operator* (const Eigen::EigenBase< Derived > &lhs, const ExtendedFloatEigenBase< EFType > &rhs) |
| template<int R, int C> | |
| ExtendedFloatArray< R, C > | operator* (const ExtendedFloatArray< R, C > &lhs, const ExtendedFloatArrayWrapper< R, C > &rhs) |
| Dimension< TransitionFunction > | transitionFunctionDimension (Eigen::Index nbState) |
| MatrixDimension | conditionalLikelihoodDimension (Eigen::Index nbState, Eigen::Index nbSite) |
| MatrixDimension | transitionMatrixDimension (std::size_t nbState) |
| RowVectorDimension | equilibriumFrequenciesDimension (std::size_t nbState) |
| std::unordered_map< std::string, std::shared_ptr< ConfiguredParameter > > | createParameterMap (Context &c, const ParameterAliasable ¶metrizable) |
| std::unordered_map< std::string, std::shared_ptr< ConfiguredParameter > > | createParameterMap (Context &c, const Parametrizable ¶metrizable) |
| NodeRefVec | createDependencyVector (const Parametrizable ¶metrizable, const std::function< NodeRef(const std::string &)> ¶meter) |
| NodeRefVec | createDependencyVector (const ParameterAliasable ¶metrizable, const std::function< NodeRef(const std::string &)> ¶meter) |
| NodeRefVec | createDependencyVector (const Parametrizable ¶metrizable, const std::function< NodeRef(const std::string &)> &getParameter) |
| Create a dependency vector suitable for a parametrizable class constructor. The vector is built from parameter names, and an opaque accessor function. For each named parameter, parameter(name) should return a valid node. If no node is found (NodeRef was null), an exception is thrown. Returned nodes must be Value<double> nodes. More... | |
| NodeRefVec | createDependencyVector (const ParameterAliasable ¶metrizable, const std::function< NodeRef(const std::string &)> &getParameter) |
| ParametrizableCollection< ConfiguredModel > | makeConfiguredModelCollection (Context &context, const SubstitutionProcessInterface &process, ParameterList &parList) |
| Make a Collection of ConfiguredModel, from the models described in the SubstitutionProcess, and sharing ConfiguredParameters from a ParameterList. More... | |
Error functions (generate a message and throw exceptions). | |
| void | failureComputeWasCalled (const std::type_info &nodeType) |
| void | failureNodeConversion (const std::type_info &handleType, const Node_DF &node) |
| void | failureDependencyNumberMismatch (const std::type_info &contextNodeType, std::size_t expectedSize, std::size_t givenSize) |
| void | failureEmptyDependency (const std::type_info &contextNodeType, std::size_t depIndex) |
| void | failureDependencyTypeMismatch (const std::type_info &contextNodeType, std::size_t depIndex, const std::type_info &expectedType, const std::type_info &givenNodeType) |
Basic dependency check primitives | |
Checks the size of a dependency vector, throws if mismatch. | |
| void | checkDependencyVectorSize (const std::type_info &contextNodeType, const NodeRefVec &deps, std::size_t expectedSize) |
| void | checkDependencyVectorMinSize (const std::type_info &contextNodeType, const NodeRefVec &deps, std::size_t expectedMinSize) |
| Checks the minimum size of a dependency vector, throws if mismatch. More... | |
| void | checkDependenciesNotNull (const std::type_info &contextNodeType, const NodeRefVec &deps) |
| Checks that all dependencies are not null, throws if not. More... | |
| void | checkNthDependencyNotNull (const std::type_info &contextNodeType, const NodeRefVec &deps, std::size_t index) |
| template<typename T > | |
| void | checkNthDependencyIs (const std::type_info &contextNodeType, const NodeRefVec &deps, std::size_t index) |
| Checks that deps[index] is a T node, throws if not. More... | |
| template<typename T1 , typename T2 > | |
| void | checkNthDependencyIs (const std::type_info &contextNodeType, const NodeRefVec &deps, std::size_t index) |
| Checks that deps[index] is a T1 or a T2 node, throws if not. More... | |
| template<typename T > | |
| void | checkDependencyRangeIs (const std::type_info &contextNodeType, const NodeRefVec &deps, std::size_t start, std::size_t end) |
| Check that deps[start, end[ contains T, throws if not. More... | |
| template<typename T > | |
| void | checkNthDependencyIsValue (const std::type_info &contextNodeType, const NodeRefVec &deps, std::size_t index) |
| Checks that deps[index] is a Value<T> node, throws if not. More... | |
| template<typename T > | |
| void | checkDependencyRangeIsValue (const std::type_info &contextNodeType, const NodeRefVec &deps, std::size_t start, std::size_t end) |
| Check that deps[start, end[ contains Value<T> nodes, throws if not. More... | |
Variables | |
| NumericConstant< char > | NodeX ('X') |
Detailed Description
Defines the basic types of data flow nodes.
POMO-like Model, based on AllelicAlphabet.
In the alphabet from which the AllelicAlphabet is built, this model needs a given mutation model with generator (aka SubstitutionModel in Bio++), and allele selection coefficients from a finesse vector (aka FrequencySet in Bio++).
If we denote ![]() this fitness, the drift term towards the fixation of allele
this fitness, the drift term towards the fixation of allele ![]() is proportional
is proportional ![]() , and follows the Moran law.
, and follows the Moran law.
We set the generator ![]() of the model on set of states
of the model on set of states ![]() ,\ with
,\ with ![]() the reversible mutation rates, with
the reversible mutation rates, with ![]() the equilibrium distribution of this mutation model, and
the equilibrium distribution of this mutation model, and ![]() , and
, and ![]() the fitnesses of the states:
the fitnesses of the states:
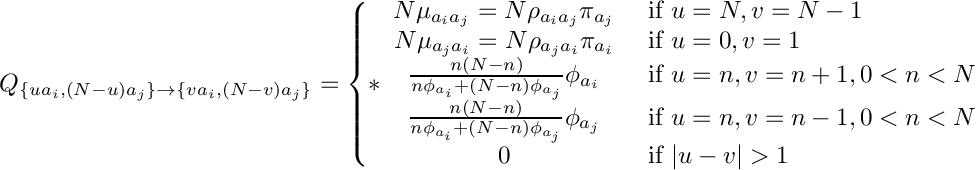
(with ![]() )
)
The equilibrium distribution ![]() is:
is:
![]()
with ![]()
The generator should be normalized to ensure one substitution per unit of time. See document online:
https://www.overleaf.com/project/639760f5aacdcfbde3ced551
See:
De Maio, N., Schrempf, D., and Kosiol, C. (2015). PoMo: An allele frequency-based approach for 279 species tree estimation. Systematic Biology, 64(6):1018–1031.
Borges R, Szöllősi GJ, Kosiol C. Quantifying GC-Biased Gene Conversion in Great Ape Genomes Using Polymorphism-Aware Models. Genetics. 2019 Aug;212(4):1321-1336.
Borges, R. and Kosiol, C. (2020). Consistency and identifiability of the polymorphism-aware phylo-genetic models. Journal of Theoretical Biology, 486:110074.
Typedef Documentation
◆ AllRatesSiteLikelihoods
Definition at line 55 of file LikelihoodCalculationOnABranch.h.
◆ BackwardLikelihoodAbove
| typedef Value< MatrixLik > bpp::BackwardLikelihoodAbove |
Definition at line 74 of file BackwardLikelihoodTree.h.
◆ BackwardLikelihoodAboveRef
| typedef ValueRef< MatrixLik > bpp::BackwardLikelihoodAboveRef |
Definition at line 75 of file BackwardLikelihoodTree.h.
◆ BackwardProportion
| using bpp::BackwardProportion = typedef CWiseMul<MatrixLik, std::tuple<double, MatrixLik> > |
: Above each node : in case of mixture of above edges
- backwardlikelihood: Matrix(state, site).
- proportion: Double
- conditionalLikelihood: Matrix(state, site).
f(State, site) = c(fromState, site) * prop
Definition at line 67 of file BackwardLikelihoodTree.h.
◆ BackwardTransition
| using bpp::BackwardTransition = typedef MatrixProduct<MatrixLik, Transposed<Eigen::MatrixXd>, MatrixLik> |
: Above each node : bottom of an edge in case of transition from upper
backwardLikelihood = f(transitionMatrix, conditionalLikelihood).
- backwardLikelihood: Matrix(state, site).
- transitionMatrix: Matrix (FromState, toState)
- conditionalLikelihood: Matrix(state, site).
f(fromState, site) = sum_toState P(fromState, toState) * c(toState, site). Using matrix multiply: f = transposed(transitionMatrix) * c.
Definition at line 55 of file BackwardLikelihoodTree.h.
◆ BaseTree
| using bpp::BaseTree = typedef AssociationTreeGlobalGraphObserver<ProcessComputationNode, ProcessComputationEdge> |
Definition at line 156 of file ProcessComputationTree.h.
◆ Bitset
| typedef std::bitset<21> bpp::Bitset |
Definition at line 20 of file DRTreeParsimonyData.h.
◆ BuildConditionalLikelihood
| typedef CWiseMul< MatrixLik, std::tuple< MatrixLik, MatrixLik > > bpp::BuildConditionalLikelihood |
Conditionallikelihood = AboveConditionalLikelihood * BelowConditionalLikelihood.
lik(state, site) = above(state, site) * below(state,site) Using member wise multiply
Definition at line 41 of file LikelihoodCalculationOnABranch.h.
◆ ConditionalLikelihood
| typedef Value< MatrixLik > bpp::ConditionalLikelihood |
Definition at line 70 of file BackwardLikelihoodTree.h.
◆ ConditionalLikelihoodDAG
| using bpp::ConditionalLikelihoodDAG = typedef AssociationDAGlobalGraphObserver<ConditionalLikelihood, uint> |
Definition at line 102 of file LikelihoodCalculationSingleProcess.h.
◆ ConditionalLikelihoodForward
| using bpp::ConditionalLikelihoodForward = typedef Value<MatrixLik> |
Interface LikelihoodTree data structure.
Stores all the inner computations:
- conditional likelihoods for each node,
- correspondence between sites in the dataset and array indices.
The structure is initiated according to a tree topology, and data can be retrieved through node ids.
- See also
- LikelihoodNode
Definition at line 89 of file ForwardLikelihoodTree.h.
◆ ConditionalLikelihoodForwardRef
| using bpp::ConditionalLikelihoodForwardRef = typedef ValueRef<MatrixLik> |
Definition at line 90 of file ForwardLikelihoodTree.h.
◆ ConditionalLikelihoodRef
| typedef ValueRef< MatrixLik > bpp::ConditionalLikelihoodRef |
Definition at line 71 of file BackwardLikelihoodTree.h.
◆ ConditionalLikelihoodTree
| using bpp::ConditionalLikelihoodTree = typedef AssociationTreeGlobalGraphObserver<ConditionalLikelihood, uint> |
Definition at line 104 of file LikelihoodCalculationSingleProcess.h.
◆ DAGindexes
| typedef std::vector< uint > bpp::DAGindexes |
Helper: create a map with mutable dataflow nodes for each branch of the tree. The map is indexed by branch ids.
Definition at line 96 of file ForwardLikelihoodTree.h.
◆ DAProb
| using bpp::DAProb = typedef AssociationDAGlobalGraphObserver<Proba, Proba> |
Definition at line 240 of file ForwardLikelihoodTree.h.
◆ DataLik
| typedef ExtendedFloat bpp::DataLik |
Definition at line 16 of file Definitions.h.
◆ EFArray
| using bpp::EFArray = typedef Eigen::Array<double, R, C> |
Definition at line 58 of file ExtendedFloatEigen.h.
◆ EFMatrix
| using bpp::EFMatrix = typedef Eigen::Matrix<double, R, C> |
Definition at line 55 of file ExtendedFloatEigen.h.
◆ EFoutput
| using bpp::EFoutput = typedef typename std::enable_if<(std::is_same<eVector, ExtendedFloatRowVectorXd>::value || std::is_same<eVector, ExtendedFloatVectorXd>::value), bool>::type |
Definition at line 86 of file DataFlowNumeric.h.
◆ EFVector
| using bpp::EFVector = typedef typename std::enable_if<(std::is_same<eVector, ExtendedFloatRowVectorXd>::value || std::is_same<eVector, ExtendedFloatVectorXd>::value), bool>::type |
Definition at line 60 of file DataFlowNumeric.h.
◆ EmissionLogk
| using bpp::EmissionLogk = typedef CWiseCompound<MatrixLik, ReductionOf<RowLik> > |
Definition at line 16 of file HmmPhyloEmissionProbabilities.h.
◆ ExtendedFloatArray
| using bpp::ExtendedFloatArray = typedef ExtendedFloatEigen<R, C, EFArray> |
Definition at line 882 of file ExtendedFloatEigen.h.
◆ ExtendedFloatArrayXd
| typedef ExtendedFloatArray<Eigen::Dynamic, 1> bpp::ExtendedFloatArrayXd |
Definition at line 896 of file ExtendedFloatEigen.h.
◆ ExtendedFloatMatrix
| using bpp::ExtendedFloatMatrix = typedef ExtendedFloatEigen<R, C, EFMatrix> |
Definition at line 879 of file ExtendedFloatEigen.h.
◆ ExtendedFloatMatrixXd
| typedef ExtendedFloatMatrix<Eigen::Dynamic, Eigen::Dynamic> bpp::ExtendedFloatMatrixXd |
Definition at line 890 of file ExtendedFloatEigen.h.
◆ ExtendedFloatRowVector
| using bpp::ExtendedFloatRowVector = typedef ExtendedFloatMatrix<1, C> |
Definition at line 888 of file ExtendedFloatEigen.h.
◆ ExtendedFloatRowVectorXd
| typedef ExtendedFloatRowVector<Eigen::Dynamic> bpp::ExtendedFloatRowVectorXd |
Definition at line 892 of file ExtendedFloatEigen.h.
◆ ExtendedFloatVector
| using bpp::ExtendedFloatVector = typedef ExtendedFloatMatrix<R, 1> |
Definition at line 885 of file ExtendedFloatEigen.h.
◆ ExtendedFloatVectorXd
| typedef ExtendedFloatVector<Eigen::Dynamic> bpp::ExtendedFloatVectorXd |
Definition at line 894 of file ExtendedFloatEigen.h.
◆ ForConvert
| using bpp::ForConvert = typedef typename std::enable_if< ((std::is_same<bVector, Vdouble>::value || std::is_same<bVector, std::vector<ExtendedFloat> >::value) && (std::is_same<eVector, Eigen::RowVectorXd>::value || std::is_same<eVector, Eigen::VectorXd>::value)), bool>::type |
Definition at line 33 of file DataFlowNumeric.h.
◆ ForConvert2
| using bpp::ForConvert2 = typedef typename std::enable_if< ((std::is_same<bVector, Vdouble>::value || std::is_same<bVector, std::vector<ExtendedFloat> >::value) && (std::is_same<eVector, Eigen::RowVectorXd>::value || std::is_same<eVector, Eigen::VectorXd>::value || std::is_same<eVector, ExtendedFloatVectorXd>::value || std::is_same<eVector, ExtendedFloatRowVectorXd>::value)), bool>::type |
Definition at line 170 of file DataFlowNumeric.h.
◆ ForwardLikelihoodBelow
| typedef Value< MatrixLik > bpp::ForwardLikelihoodBelow |
Definition at line 93 of file ForwardLikelihoodTree.h.
◆ ForwardLikelihoodBelowRef
| typedef ValueRef< MatrixLik > bpp::ForwardLikelihoodBelowRef |
Definition at line 94 of file ForwardLikelihoodTree.h.
◆ ForwardProportion
| using bpp::ForwardProportion = typedef CWiseMul<MatrixLik, std::tuple<double, MatrixLik> > |
forwardLikelihood = f(transitionMatrix, proportion).
- forwardLikelihood: Matrix(state, site).
- proportion: Double
- conditionalLikelihood: Matrix(state, site).
f(State, site) = c(fromState, site) * prop
Definition at line 72 of file ForwardLikelihoodTree.h.
◆ ForwardTransition
| typedef MatrixProduct< MatrixLik, Eigen::MatrixXd, MatrixLik > bpp::ForwardTransition |
forwardLikelihood = f(transitionMatrix, conditionalLikelihood).
- forwardLikelihood: Matrix(state, site).
- transitionMatrix: Matrix (fromState, toState)
- conditionalLikelihood: Matrix(state, site).
f(toState, site) = sum_fromState P(fromState, toState) * c(fromState, site).
Definition at line 58 of file ForwardLikelihoodTree.h.
◆ ForwardTransitionFunction
| using bpp::ForwardTransitionFunction = typedef CWiseApply<MatrixLik, MatrixLik, TransitionFunction> |
Definition at line 61 of file ForwardLikelihoodTree.h.
◆ INode
| typedef NodeTemplate<TreeDrawingNodeInfo> bpp::INode |
Definition at line 43 of file AbstractTreeDrawing.h.
◆ LikelihoodFromRootConditional
| using bpp::LikelihoodFromRootConditional = typedef MatrixProduct<RowLik, RowLik, MatrixLik> |
likelihood = f(equilibriumFrequencies, rootConditionalLikelihood).
- likelihood: RowVector(site).
- equilibriumFrequencies: RowVector(state).
- rootConditionalLikelihood: Matrix(state, site).
likelihood(site) = sum_state equFreqs(state) * rootConditionalLikelihood(state, site). Using matrix multiply: likelihood = equilibriumFrequencies * rootConditionalLikelihood.
Definition at line 63 of file LikelihoodCalculationSingleProcess.h.
◆ LikelihoodFromRootConditionalAtRoot
| typedef MatrixProduct< RowLik, Eigen::RowVectorXd, MatrixLik > bpp::LikelihoodFromRootConditionalAtRoot |
Likelihood computed on a branch. This likelihood is dependent on a LikelihoodCalculationSingleProcess, from which it will get the conditional likelihoods at both ends of the branch, a Branch Node (with the parameterized length), as well as a Model DF.
Definition at line 25 of file LikelihoodCalculationOnABranch.h.
◆ MatchingType
| typedef Eigen::Matrix<size_t, -1, 2> bpp::MatchingType |
Definition at line 290 of file DataFlowCWise.h.
◆ MatrixLik
| typedef ExtendedFloatMatrixXd bpp::MatrixLik |
Definition at line 13 of file Definitions.h.
◆ MixtureBackward
| using bpp::MixtureBackward = typedef CWiseAdd<MatrixLik, ReductionOf<MatrixLik> > |
: At the top of each edge below a mixture node
backwardLikelihood = f(backwardLikelihood[father[i]] for i) backwardLikelihood: Matrix(state, site). backwardLikelihood[i]: Matrix(state, site).
c(state, site) = sum_i f_i(state, site) Using member wise addition: c = sum_member_i f_i Using member wise multiplication: c(state, site) = prod_i f_i(state, site).
Definition at line 41 of file BackwardLikelihoodTree.h.
◆ MixtureForward
| using bpp::MixtureForward = typedef CWiseAdd<MatrixLik, ReductionOf<MatrixLik> > |
conditionalLikelihood = f(forwardLikelihood[children[i]] for i). conditionalLikelihood: Matrix(state, site). forwardLikelihood[i]: Matrix(state, site).
c(state, site) = sum_i f_i(state, site) Using member wise addition: c = sum_member_i f_i
Definition at line 48 of file ForwardLikelihoodTree.h.
◆ NodeRef
| using bpp::NodeRef = typedef std::shared_ptr<Node_DF> |
Node instances are always manipulated as shared pointers: provide a short alias.
Definition at line 78 of file DataFlow.h.
◆ NodeRefVec
| using bpp::NodeRefVec = typedef std::vector<NodeRef> |
Alias for a dependency vector (of NodeRef).
Definition at line 81 of file DataFlow.h.
◆ PatternType
| typedef Eigen::Matrix<size_t, -1, 1> bpp::PatternType |
Definition at line 181 of file DataFlowCWise.h.
◆ Proba
| using bpp::Proba = typedef Value<double> |
DAG with the same shape as ForwardLikelihoodTree with computations of probabilities of nodes & branches.
Definition at line 237 of file ForwardLikelihoodTree.h.
◆ ProbaMul
| using bpp::ProbaMul = typedef CWiseMul<double, std::tuple<double, double> > |
Definition at line 242 of file ForwardLikelihoodTree.h.
◆ ProbaRef
| using bpp::ProbaRef = typedef std::shared_ptr<Value<double> > |
Definition at line 238 of file ForwardLikelihoodTree.h.
◆ ProbaSum
| using bpp::ProbaSum = typedef CWiseAdd<double, ReductionOf<double> > |
Definition at line 244 of file ForwardLikelihoodTree.h.
◆ ProcessComputationNodeRef
| using bpp::ProcessComputationNodeRef = typedef std::shared_ptr<ProcessComputationNode> |
Definition at line 71 of file ProcessComputationTree.h.
◆ ProcessEdgePtr
| using bpp::ProcessEdgePtr = typedef std::shared_ptr<ProcessEdge> |
Definition at line 147 of file ProcessTree.h.
◆ ProcessNode
Definition at line 145 of file ProcessTree.h.
◆ ProcessNodePtr
| using bpp::ProcessNodePtr = typedef std::shared_ptr<ProcessNode> |
Definition at line 148 of file ProcessTree.h.
◆ RowLik
| typedef ExtendedFloatRowVectorXd bpp::RowLik |
Definition at line 14 of file Definitions.h.
◆ SiteLikelihoods
| typedef Value< RowLik > bpp::SiteLikelihoods |
Definition at line 121 of file LikelihoodCalculation.h.
◆ SiteLikelihoodsDAG
| using bpp::SiteLikelihoodsDAG = typedef AssociationDAGlobalGraphObserver<SiteLikelihoods, uint> |
Definition at line 106 of file LikelihoodCalculationSingleProcess.h.
◆ SiteLikelihoodsRef
| typedef ValueRef< RowLik > bpp::SiteLikelihoodsRef |
Definition at line 122 of file LikelihoodCalculation.h.
◆ SiteLikelihoodsTree
| using bpp::SiteLikelihoodsTree = typedef AssociationTreeGlobalGraphObserver<SiteLikelihoods, uint> |
Definition at line 108 of file LikelihoodCalculationSingleProcess.h.
◆ SiteWeights
| typedef NumericConstant< Eigen::RowVectorXi > bpp::SiteWeights |
Definition at line 57 of file LikelihoodCalculationOnABranch.h.
◆ SNode
| typedef NodeTemplate<SimData> bpp::SNode |
Definition at line 52 of file NonHomogeneousSequenceSimulator.h.
◆ SpeciationBackward
| using bpp::SpeciationBackward = typedef CWiseMul<MatrixLik, ReductionOf<MatrixLik> > |
: At the top of each edge below a speciation node
conditionalLikelihood = f(backwardLikelihood[father[i]], forwardlikelihood[brothers[i]]). conditionalLikelihood: Matrix(state, site). backwardLikelihood, forwardlikelihood: Matrix(state, site).
Using member wise multiplication: c(state, site) = prod_i f_i(state, site).
Definition at line 28 of file BackwardLikelihoodTree.h.
◆ SpeciationForward
| using bpp::SpeciationForward = typedef CWiseMul<MatrixLik, ReductionOf<MatrixLik> > |
conditionalLikelihood = f(forwardLikelihood[children[i]] for i). conditionalLikelihood: Matrix(state, site). forwardLikelihood[i]: Matrix(state, site).
c(state, site) = prod_i f_i(state, site). Using member wise multiply: c = prod_member_i f_i.
Definition at line 38 of file ForwardLikelihoodTree.h.
◆ Speciesindex
| typedef uint bpp::Speciesindex |
Definition at line 97 of file ForwardLikelihoodTree.h.
◆ SPTree
Definition at line 63 of file SimpleSubstitutionProcessSiteSimulator.h.
◆ TotalLogLikelihood
| typedef SumOfLogarithms< RowLik > bpp::TotalLogLikelihood |
totalLikelihood = product_site likelihood(site).
- likelihood: RowVector (site).
- totalLikelihood: Extended float.
Definition at line 33 of file LikelihoodCalculationOnABranch.h.
◆ TransitionFunction
| using bpp::TransitionFunction = typedef std::function<VectorLik (const VectorLik&)> |
Definition at line 198 of file DataFlowNumeric.h.
◆ ValueRef
| using bpp::ValueRef = typedef std::shared_ptr<Value<T> > |
Shared pointer alias for Value<T>.
Definition at line 84 of file DataFlow.h.
◆ VDataLik
| typedef std::vector<DataLik> bpp::VDataLik |
Definition at line 23 of file Definitions.h.
◆ VectorLik
| typedef ExtendedFloatVectorXd bpp::VectorLik |
Definition at line 15 of file Definitions.h.
◆ VVDataLik
| typedef std::vector<VDataLik> bpp::VVDataLik |
Definition at line 24 of file Definitions.h.
Enumeration Type Documentation
◆ DotOptions
|
strong |
Dot output Flags to control dot output of dataflow graphs.
| Enumerator | |
|---|---|
| None | |
| DetailedNodeInfo | |
| FollowUpwardLinks | |
| ShowDependencyIndex | |
Definition at line 46 of file DataFlow.h.
◆ NumericalDerivativeType
|
strong |
| Enumerator | |
|---|---|
| Disabled | |
| ThreePoints | |
| FivePoints | |
Definition at line 2700 of file DataFlowCWiseComputing.h.
◆ NumericalProperty
|
strong |
Numerical properties for DF Node.
| Enumerator | |
|---|---|
| Constant | |
| ConstantZero | |
| ConstantOne | |
| ConstantIdentity | |
Definition at line 87 of file DataFlow.h.
Function Documentation
◆ abs()
|
inline |
Definition at line 479 of file ExtendedFloat.h.
References bpp::ExtendedFloat::abs().
Referenced by bpp::ExtendedFloat::abs(), bpp::HierarchicalClustering::computeDistancesFromPair(), bpp::AbstractDFPSubstitutionModel::getCodonsMulRate(), bpp::AbstractSubstitutionModel::getPij_t(), bpp::TsTvSubstitutionRegister::getType(), bpp::DecompositionMethods::jFunction_(), bpp::ExtendedFloat::normalize_big(), bpp::ExtendedFloat::normalize_small(), bpp::OptimizationTools::OptimizationOptions::OptimizationOptions(), bpp::LegacyPhylogeneticsApplicationTools::optimizeParameters(), bpp::UniformizationSubstitutionCount::setSubstitutionModel(), bpp::SimpleMutationProcess::SimpleMutationProcess(), bpp::UniformizationSubstitutionCount::UniformizationSubstitutionCount(), bpp::AbstractSubstitutionModel::updateMatrices_(), bpp::gBGC::updateMatrices_(), bpp::RN95::updateMatrices_(), bpp::RN95s::updateMatrices_(), bpp::POMO::updateMatrices_(), and bpp::YpR::updateMatrices_().
◆ accessValueConstCast()
| const T & bpp::accessValueConstCast | ( | const Node_DF & | node | ) |
Helper: access value of Node as a Value<T> with unchecked cast.
Definition at line 424 of file DataFlow.h.
◆ cachedAs() [1/2]
| std::shared_ptr< T > bpp::cachedAs | ( | Context & | c, |
| std::shared_ptr< T > && | newNode | ||
| ) |
Helper: Same as Context::cached but with a shared_ptr<T> node.
Definition at line 611 of file DataFlow.h.
References bpp::Context::cached().
◆ cachedAs() [2/2]
| std::shared_ptr< T > bpp::cachedAs | ( | Context & | c, |
| std::shared_ptr< T > & | newNode | ||
| ) |
Definition at line 617 of file DataFlow.h.
References bpp::Context::cached().
◆ checkDependenciesNotNull()
| void bpp::checkDependenciesNotNull | ( | const std::type_info & | contextNodeType, |
| const NodeRefVec & | deps | ||
| ) |
Checks that all dependencies are not null, throws if not.
Definition at line 103 of file DataFlow.cpp.
References failureEmptyDependency().
Referenced by bpp::ScalarProduct< R, T0, T1 >::create(), bpp::ProbabilitiesFromDiscreteDistribution::create(), bpp::ProbabilitiesFromMixedModel::create(), bpp::ValueFromConfiguredParameter::create(), bpp::CondLikelihood::create(), bpp::ForwardHmmLikelihood_DF::create(), bpp::ForwardHmmDLikelihood_DF::create(), bpp::ForwardHmmD2Likelihood_DF::create(), bpp::BackwardHmmLikelihood_DF::create(), bpp::SumOfLogarithms< F >::create(), bpp::CWiseFill< R, T >::create(), bpp::CWisePattern< R >::create(), bpp::CWiseMatching< R, ReductionOf< T > >::create(), bpp::CWiseCompound< R, ReductionOf< T > >::create(), bpp::CWiseApply< R, T, F >::create(), bpp::CWiseAdd< R, std::tuple< T0, T1 > >::create(), bpp::CWiseSub< R, std::tuple< T0, T1 > >::create(), bpp::CWiseAdd< R, ReductionOf< T > >::create(), bpp::CWiseAdd< R, T >::create(), bpp::CWiseMean< R, ReductionOf< T >, ReductionOf< P > >::create(), bpp::CWiseMul< R, std::tuple< T0, T1 > >::create(), bpp::CWiseMul< R, ReductionOf< T > >::create(), bpp::CWiseDiv< R, std::tuple< T0, T1 > >::create(), bpp::MatrixProduct< R, T0, T1 >::create(), bpp::Convert< R, F >::create(), bpp::Identity< R >::create(), bpp::CWiseNegate< T >::create(), bpp::CWiseInverse< T >::create(), bpp::CWiseLog< T >::create(), bpp::CWiseExp< T >::create(), bpp::TransitionFunctionFromModel::create(), bpp::LogSumExp< R, T0, T1 >::create(), bpp::ConfiguredParameter::create(), bpp::CWiseConstantPow< T >::create(), bpp::ShiftDelta< T >::create(), bpp::CombineDeltaShifted< T >::create(), bpp::ShiftParameter::create(), bpp::ProbabilityFromMixedModel::create(), bpp::ProbabilityFromDiscreteDistribution::create(), bpp::CategoryFromDiscreteDistribution::create(), bpp::ConfiguredParametrizable::createConfigured(), bpp::ConfiguredParametrizable::createDouble(), and bpp::ConfiguredParameter::resetDependencies().
◆ checkDependencyRangeIs()
| void bpp::checkDependencyRangeIs | ( | const std::type_info & | contextNodeType, |
| const NodeRefVec & | deps, | ||
| std::size_t | start, | ||
| std::size_t | end | ||
| ) |
Check that deps[start, end[ contains T, throws if not.
Definition at line 473 of file DataFlow.h.
◆ checkDependencyRangeIsValue()
| void bpp::checkDependencyRangeIsValue | ( | const std::type_info & | contextNodeType, |
| const NodeRefVec & | deps, | ||
| std::size_t | start, | ||
| std::size_t | end | ||
| ) |
Check that deps[start, end[ contains Value<T> nodes, throws if not.
Definition at line 492 of file DataFlow.h.
◆ checkDependencyVectorMinSize()
| void bpp::checkDependencyVectorMinSize | ( | const std::type_info & | contextNodeType, |
| const NodeRefVec & | deps, | ||
| std::size_t | expectedMinSize | ||
| ) |
Checks the minimum size of a dependency vector, throws if mismatch.
Definition at line 93 of file DataFlow.cpp.
References failureDependencyNumberMismatch().
Referenced by bpp::SumOfLogarithms< F >::create(), bpp::ConfiguredParametrizable::createMatrix(), bpp::ConfiguredParametrizable::createRowVector(), and bpp::ConfiguredParametrizable::createVector().
◆ checkDependencyVectorSize()
| void bpp::checkDependencyVectorSize | ( | const std::type_info & | contextNodeType, |
| const NodeRefVec & | deps, | ||
| std::size_t | expectedSize | ||
| ) |
Definition at line 83 of file DataFlow.cpp.
References failureDependencyNumberMismatch().
Referenced by bpp::ScalarProduct< R, T0, T1 >::create(), bpp::ProbabilitiesFromDiscreteDistribution::create(), bpp::ProbabilitiesFromMixedModel::create(), bpp::ValueFromConfiguredParameter::create(), bpp::CondLikelihood::create(), bpp::ForwardHmmLikelihood_DF::create(), bpp::ForwardHmmDLikelihood_DF::create(), bpp::ForwardHmmD2Likelihood_DF::create(), bpp::BackwardHmmLikelihood_DF::create(), bpp::CWiseFill< R, T >::create(), bpp::CWisePattern< R >::create(), bpp::CWiseApply< R, T, F >::create(), bpp::CWiseAdd< R, std::tuple< T0, T1 > >::create(), bpp::CWiseSub< R, std::tuple< T0, T1 > >::create(), bpp::CWiseAdd< R, T >::create(), bpp::CWiseMul< R, std::tuple< T0, T1 > >::create(), bpp::CWiseDiv< R, std::tuple< T0, T1 > >::create(), bpp::MatrixProduct< R, T0, T1 >::create(), bpp::Convert< R, F >::create(), bpp::Identity< R >::create(), bpp::CWiseNegate< T >::create(), bpp::CWiseInverse< T >::create(), bpp::CWiseLog< T >::create(), bpp::CWiseExp< T >::create(), bpp::TransitionFunctionFromModel::create(), bpp::LogSumExp< R, T0, T1 >::create(), bpp::ConfiguredParameter::create(), bpp::CWiseConstantPow< T >::create(), bpp::ShiftDelta< T >::create(), bpp::CombineDeltaShifted< T >::create(), bpp::ShiftParameter::create(), bpp::ProbabilityFromMixedModel::create(), bpp::ProbabilityFromDiscreteDistribution::create(), bpp::CategoryFromDiscreteDistribution::create(), bpp::ConfiguredParametrizable::createConfigured(), bpp::ConfiguredParametrizable::createDouble(), and bpp::ConfiguredParameter::resetDependencies().
◆ checkNthDependencyIs() [1/2]
| void bpp::checkNthDependencyIs | ( | const std::type_info & | contextNodeType, |
| const NodeRefVec & | deps, | ||
| std::size_t | index | ||
| ) |
Checks that deps[index] is a T node, throws if not.
Definition at line 447 of file DataFlow.h.
References failureDependencyTypeMismatch().
◆ checkNthDependencyIs() [2/2]
| void bpp::checkNthDependencyIs | ( | const std::type_info & | contextNodeType, |
| const NodeRefVec & | deps, | ||
| std::size_t | index | ||
| ) |
Checks that deps[index] is a T1 or a T2 node, throws if not.
Definition at line 457 of file DataFlow.h.
References prettyTypeName(), and to_string().
◆ checkNthDependencyIsValue()
| void bpp::checkNthDependencyIsValue | ( | const std::type_info & | contextNodeType, |
| const NodeRefVec & | deps, | ||
| std::size_t | index | ||
| ) |
Checks that deps[index] is a Value<T> node, throws if not.
Definition at line 484 of file DataFlow.h.
◆ checkNthDependencyNotNull()
| void bpp::checkNthDependencyNotNull | ( | const std::type_info & | contextNodeType, |
| const NodeRefVec & | deps, | ||
| std::size_t | index | ||
| ) |
Definition at line 114 of file DataFlow.cpp.
References failureEmptyDependency().
Referenced by bpp::ConfiguredParametrizable::createMatrix(), bpp::ConfiguredParametrizable::createRowVector(), and bpp::ConfiguredParametrizable::createVector().
◆ checkRecreateWithoutDependencies()
| void bpp::checkRecreateWithoutDependencies | ( | const std::type_info & | contextNodeType, |
| const NodeRefVec & | deps | ||
| ) |
Definition at line 173 of file DataFlowNumeric.cpp.
References prettyTypeName().
Referenced by bpp::ConstantZero< T >::recreate(), bpp::ConstantOne< T >::recreate(), bpp::NumericMutable< T >::recreate(), bpp::NumericConstant< T >::recreate(), and bpp::Sequence_DF::recreate().
◆ combineHash()
| void bpp::combineHash | ( | std::size_t & | seed, |
| const T & | t | ||
| ) |
Combine hashable value to a hash, from Boost library.
Definition at line 33 of file DataFlow.h.
Referenced by bpp::numeric::hash(), hash(), bpp::CWiseConstantPow< T >::hashAdditionalArguments(), bpp::ShiftDelta< T >::hashAdditionalArguments(), bpp::CombineDeltaShifted< T >::hashAdditionalArguments(), bpp::ShiftParameter::hashAdditionalArguments(), and bpp::Context::CachedNodeRefHash::operator()().
◆ conditionalLikelihoodDimension()
|
inline |
Definition at line 25 of file ForwardLikelihoodTree.h.
Referenced by bpp::ForwardLikelihoodTree::initialize(), bpp::LikelihoodCalculationSingleProcess::makeLikelihoodsAtDAGNode_(), and bpp::LikelihoodCalculationSingleProcess::makeLikelihoodsAtNode_().
◆ constexpr_power()
|
constexpr |
Definition at line 18 of file ExtendedFloat.h.
References constexpr_power().
Referenced by constexpr_power().
◆ convert()
|
inline |
Definition at line 486 of file ExtendedFloat.h.
References bpp::ExtendedFloat::exponent_part(), bpp::ExtendedFloat::float_part(), and bpp::ExtendedFloat::radix.
Referenced by bpp::CWiseFill< R, T >::compute(), bpp::Convert< R, F >::compute(), bpp::ForwardHmmLikelihood_DF::compute(), bpp::ForwardHmmDLikelihood_DF::compute(), bpp::ForwardHmmD2Likelihood_DF::compute(), bpp::BackwardHmmLikelihood_DF::compute(), bpp::SubstitutionMappingTools::computeCounts(), bpp::SubstitutionMappingTools::computeNormalizations(), bpp::RewardMappingTools::computeRewardVectors(), copyBppToEigen(), copyEigenToBpp(), bpp::AbstractPhyloLikelihood::getFirstOrderDerivative(), bpp::MultiProcessSequencePhyloLikelihood::getLikelihoodPerSitePerProcess(), bpp::AbstractAlignedPhyloLikelihood::getLogLikelihoodForASite(), bpp::OneProcessSequencePhyloLikelihood::getPosteriorProbabilitiesForSitePerClass(), bpp::SingleProcessPhyloLikelihood::getPosteriorProbabilitiesForSitePerClass(), bpp::MixtureProcessPhyloLikelihood::getPosteriorProbabilitiesPerSitePerProcess(), bpp::OneProcessSequencePhyloLikelihood::getPosteriorRatePerSite(), bpp::SingleProcessPhyloLikelihood::getPosteriorRatePerSite(), bpp::AbstractPhyloLikelihood::getSecondOrderDerivative(), bpp::AbstractPhyloLikelihood::getValue(), bpp::GivenDataSubstitutionProcessSiteSimulator::init(), bpp::OptimizationTools::OptimizationOptions::OptimizationOptions(), and bpp::LegacyPhylogeneticsApplicationTools::optimizeParameters().
◆ convertRef()
| std::shared_ptr< T > bpp::convertRef | ( | const std::shared_ptr< U > & | from | ) |
Convert a node ref with runtime type check.
Definition at line 320 of file DataFlow.h.
References failureNodeConversion().
◆ copyBppToEigen() [1/13]
| void bpp::copyBppToEigen | ( | const bpp::Matrix< double > & | bppMatrix, |
| Eigen::MatrixXd & | eigenMatrix | ||
| ) |
Definition at line 87 of file DataFlowNumeric.cpp.
References bpp::Matrix< class >::getNumberOfColumns(), and bpp::Matrix< class >::getNumberOfRows().
◆ copyBppToEigen() [2/13]
| void bpp::copyBppToEigen | ( | const bpp::Matrix< double > & | bppMatrix, |
| ExtendedFloatMatrixXd & | eigenMatrix | ||
| ) |
Definition at line 65 of file DataFlowNumeric.cpp.
References bpp::ExtendedFloatEigen< R, C, EigenType >::fill(), bpp::ExtendedFloatEigen< R, C, EigenType >::float_part(), bpp::Matrix< class >::getNumberOfColumns(), bpp::Matrix< class >::getNumberOfRows(), and bpp::ExtendedFloatEigen< R, C, EigenType >::resize().
◆ copyBppToEigen() [3/13]
| template void bpp::copyBppToEigen | ( | const bpp::Vdouble & | bppVector, |
| Eigen::RowVectorXd & | eigenVector | ||
| ) |
◆ copyBppToEigen() [4/13]
| template void bpp::copyBppToEigen | ( | const bpp::Vdouble & | bppVector, |
| Eigen::VectorXd & | eigenVector | ||
| ) |
◆ copyBppToEigen() [5/13]
| template void bpp::copyBppToEigen | ( | const bpp::Vdouble & | bppVector, |
| ExtendedFloatRowVectorXd & | eigenVector | ||
| ) |
◆ copyBppToEigen() [6/13]
| void bpp::copyBppToEigen | ( | const bVector & | bppVector, |
| eVector & | eigenVector | ||
| ) |
Definition at line 41 of file DataFlowNumeric.h.
References convert().
◆ copyBppToEigen() [7/13]
| void bpp::copyBppToEigen | ( | const std::vector< Eigen::VectorXd > & | bppVector, |
| Eigen::MatrixXd & | eigenMatrix | ||
| ) |
Definition at line 47 of file DataFlowNumeric.cpp.
◆ copyBppToEigen() [8/13]
| template void bpp::copyBppToEigen | ( | const std::vector< ExtendedFloat > & | bppVector, |
| Eigen::RowVectorXd & | eigenVector | ||
| ) |
Referenced by bpp::CWiseApply< R, T, F >::compute(), bpp::TransitionMatrixFromModel::compute(), bpp::TransitionMatrixFromTransitionMatrix::compute(), bpp::ForwardHmmLikelihood_DF::compute(), bpp::ForwardHmmDLikelihood_DF::compute(), bpp::ForwardHmmD2Likelihood_DF::compute(), bpp::BackwardHmmLikelihood_DF::compute(), copyBppToEigen(), bpp::OneProcessSequencePhyloLikelihood::getPosteriorProbabilitiesPerSitePerClass(), and bpp::SingleProcessPhyloLikelihood::getPosteriorProbabilitiesPerSitePerClass().
◆ copyBppToEigen() [9/13]
| template void bpp::copyBppToEigen | ( | const std::vector< ExtendedFloat > & | bppVector, |
| Eigen::VectorXd & | eigenVector | ||
| ) |
◆ copyBppToEigen() [10/13]
| void bpp::copyBppToEigen | ( | const std::vector< ExtendedFloat > & | bppVector, |
| eVector & | eigenVector | ||
| ) |
Definition at line 64 of file DataFlowNumeric.h.
◆ copyBppToEigen() [11/13]
| template void bpp::copyBppToEigen | ( | const std::vector< ExtendedFloat > & | bppVector, |
| ExtendedFloatRowVectorXd & | eigenVector | ||
| ) |
◆ copyBppToEigen() [12/13]
| void bpp::copyBppToEigen | ( | const std::vector< ExtendedFloatVectorXd > & | bppVector, |
| ExtendedFloatMatrixXd & | eigenVector | ||
| ) |
Definition at line 31 of file DataFlowNumeric.cpp.
References bpp::ExtendedFloatEigen< R, C, EigenType >::exponent_part(), and bpp::ExtendedFloatEigen< R, C, EigenType >::float_part().
◆ copyBppToEigen() [13/13]
| void bpp::copyBppToEigen | ( | const Vdouble & | bppVector, |
| eVector & | eigenVector | ||
| ) |
Definition at line 91 of file DataFlowNumeric.h.
References copyBppToEigen().
◆ copyEigenToBpp() [1/10]
| void bpp::copyEigenToBpp | ( | const bVector & | eigenVector, |
| eVector & | bppVector | ||
| ) |
Definition at line 181 of file DataFlowNumeric.h.
References convert().
◆ copyEigenToBpp() [2/10]
| template void bpp::copyEigenToBpp | ( | const Eigen::MatrixXd & | eigenMatrix, |
| std::vector< std::vector< bpp::ExtendedFloat > > & | bppMatrix | ||
| ) |
◆ copyEigenToBpp() [3/10]
| template void bpp::copyEigenToBpp | ( | const Eigen::MatrixXd & | eigenMatrix, |
| std::vector< std::vector< double > > & | bppMatrix | ||
| ) |
◆ copyEigenToBpp() [4/10]
| void bpp::copyEigenToBpp | ( | const Eigen::MatrixXd & | eigenMatrix, |
| std::vector< std::vector< eType > > & | bppMatrix | ||
| ) |
Definition at line 144 of file DataFlowNumeric.h.
◆ copyEigenToBpp() [5/10]
| template void bpp::copyEigenToBpp | ( | const ExtendedFloatMatrixXd & | eigenMatrix, |
| std::vector< std::vector< bpp::ExtendedFloat > > & | bppMatrix | ||
| ) |
◆ copyEigenToBpp() [6/10]
| template void bpp::copyEigenToBpp | ( | const ExtendedFloatMatrixXd & | eigenMatrix, |
| std::vector< std::vector< double > > & | bppMatrix | ||
| ) |
Referenced by bpp::MarginalAncestralReconstruction::getAncestralStatesForNode(), bpp::AlignedLikelihoodCalculation::getLikelihoodPerSite(), bpp::OnABranchPhyloLikelihood::getLikelihoodPerSitePerClass(), bpp::OneProcessSequencePhyloLikelihood::getLikelihoodPerSitePerClass(), bpp::SingleProcessPhyloLikelihood::getLikelihoodPerSitePerClass(), bpp::AlignedPhyloLikelihoodAutoCorrelation::getPosteriorProbabilitiesForASitePerAligned(), bpp::AlignedPhyloLikelihoodHmm::getPosteriorProbabilitiesForASitePerAligned(), bpp::AlignedPhyloLikelihoodAutoCorrelation::getPosteriorProbabilitiesPerSitePerAligned(), bpp::AlignedPhyloLikelihoodHmm::getPosteriorProbabilitiesPerSitePerAligned(), bpp::OneProcessSequencePhyloLikelihood::getPosteriorProbabilitiesPerSitePerClass(), bpp::SingleProcessPhyloLikelihood::getPosteriorProbabilitiesPerSitePerClass(), bpp::AutoCorrelationProcessPhyloLikelihood::getPosteriorProbabilitiesPerSitePerProcess(), bpp::HmmProcessPhyloLikelihood::getPosteriorProbabilitiesPerSitePerProcess(), bpp::OneProcessSequencePhyloLikelihood::getPosteriorStateFrequencies(), bpp::SingleProcessPhyloLikelihood::getPosteriorStateFrequencies(), bpp::GivenDataSubstitutionProcessSiteSimulator::init(), and bpp::PhylogeneticsApplicationTools::printParameters().
◆ copyEigenToBpp() [7/10]
| void bpp::copyEigenToBpp | ( | const ExtendedFloatMatrixXd & | eigenMatrix, |
| std::vector< std::vector< eType > > & | bppMatrix | ||
| ) |
Definition at line 120 of file DataFlowNumeric.h.
References bpp::ExtendedFloatEigen< R, C, EigenType >::cols(), convert(), and bpp::ExtendedFloatEigen< R, C, EigenType >::rows().
◆ copyEigenToBpp() [8/10]
| void bpp::copyEigenToBpp | ( | const MatrixLik & | eigenMatrix, |
| bpp::Matrix< double > & | bppMatrix | ||
| ) |
Definition at line 118 of file DataFlowNumeric.cpp.
References bpp::ExtendedFloatEigen< R, C, EigenType >::cols(), convert(), bpp::Matrix< class >::resize(), and bpp::ExtendedFloatEigen< R, C, EigenType >::rows().
◆ copyEigenToBpp() [9/10]
| template void bpp::copyEigenToBpp | ( | const RowLik & | eigenVector, |
| std::vector< ExtendedFloat > & | bppVector | ||
| ) |
◆ copyEigenToBpp() [10/10]
◆ createDependencyVector() [1/4]
| NodeRefVec bpp::createDependencyVector | ( | const ParameterAliasable & | parametrizable, |
| const std::function< NodeRef(const std::string &)> & | getParameter | ||
| ) |
◆ createDependencyVector() [2/4]
| NodeRefVec bpp::createDependencyVector | ( | const ParameterAliasable & | parametrizable, |
| const std::function< NodeRef(const std::string &)> & | parameter | ||
| ) |
Definition at line 66 of file Parametrizable.cpp.
References bpp::ParameterAliasable::getIndependentParameters(), and bpp::ParameterList::size().
◆ createDependencyVector() [3/4]
| NodeRefVec bpp::createDependencyVector | ( | const Parametrizable & | parametrizable, |
| const std::function< NodeRef(const std::string &)> & | getParameter | ||
| ) |
Create a dependency vector suitable for a parametrizable class constructor. The vector is built from parameter names, and an opaque accessor function. For each named parameter, parameter(name) should return a valid node. If no node is found (NodeRef was null), an exception is thrown. Returned nodes must be Value<double> nodes.
◆ createDependencyVector() [4/4]
| NodeRefVec bpp::createDependencyVector | ( | const Parametrizable & | parametrizable, |
| const std::function< NodeRef(const std::string &)> & | parameter | ||
| ) |
Definition at line 48 of file Parametrizable.cpp.
References bpp::Parametrizable::getParameters(), and bpp::ParameterList::size().
Referenced by bpp::ConfiguredParametrizable::createConfigured().
◆ createParameterMap() [1/2]
| std::unordered_map< std::string, std::shared_ptr< ConfiguredParameter > > bpp::createParameterMap | ( | Context & | c, |
| const ParameterAliasable & | parametrizable | ||
| ) |
Definition at line 17 of file Parametrizable.cpp.
References bpp::NumericMutable< T >::create(), bpp::ConfiguredParameter::create(), bpp::ParameterAliasable::getIndependentParameters(), and bpp::ParameterList::size().
Referenced by bpp::ConfiguredParametrizable::createConfigured().
◆ createParameterMap() [2/2]
| std::unordered_map< std::string, std::shared_ptr< ConfiguredParameter > > bpp::createParameterMap | ( | Context & | c, |
| const Parametrizable & | parametrizable | ||
| ) |
Helper: create a map with mutable dataflow nodes for each parameter of the parametrizable. The map is indexed by parameter names.
Definition at line 33 of file Parametrizable.cpp.
References bpp::NumericMutable< T >::create(), bpp::ConfiguredParameter::create(), bpp::Parametrizable::getParameters(), and bpp::ParameterList::size().
◆ dotIdentifier()
|
static |
Definition at line 469 of file DataFlow.cpp.
References to_string().
Referenced by writeDotEdge(), and writeDotNode().
◆ dotLabelEscape()
|
static |
Definition at line 449 of file DataFlow.cpp.
Referenced by writeDotNode().
◆ equilibriumFrequenciesDimension()
|
inline |
◆ exp() [1/2]
|
inline |
Definition at line 474 of file ExtendedFloat.h.
References bpp::ExtendedFloat::exp().
Referenced by bpp::CWiseExp< T >::compute(), bpp::LogSumExp< R, T0, T1 >::compute(), bpp::MultinomialFromTransitionModel::compute_d2Multinomial_dt2_(), bpp::MultinomialFromTransitionModel::compute_dMultinomial_dt_(), bpp::MultinomialFromTransitionModel::compute_Multinomial_(), bpp::UniformizationSubstitutionCount::computeCounts_(), bpp::PairedSiteLikelihoods::computeExpectedLikelihoodWeights(), bpp::BinarySubstitutionModel::d2Pij_dt2(), bpp::F84::d2Pij_dt2(), bpp::HKY85::d2Pij_dt2(), bpp::TwoParameterBinarySubstitutionModel::d2Pij_dt2(), bpp::EquiprobableSubstitutionModel::d2Pij_dt2(), bpp::F81::d2Pij_dt2(), bpp::JCnuc::d2Pij_dt2(), bpp::K80::d2Pij_dt2(), bpp::T92::d2Pij_dt2(), bpp::TN93::d2Pij_dt2(), bpp::JCprot::d2Pij_dt2(), bpp::RE08::d2Pij_dt2(), bpp::OneChangeTransitionModel::d2Pij_dt2(), bpp::AbstractMutationProcess::detailedEvolve(), bpp::BinarySubstitutionModel::dPij_dt(), bpp::F84::dPij_dt(), bpp::HKY85::dPij_dt(), bpp::TwoParameterBinarySubstitutionModel::dPij_dt(), bpp::EquiprobableSubstitutionModel::dPij_dt(), bpp::F81::dPij_dt(), bpp::JCnuc::dPij_dt(), bpp::K80::dPij_dt(), bpp::T92::dPij_dt(), bpp::TN93::dPij_dt(), bpp::JCprot::dPij_dt(), bpp::RE08::dPij_dt(), bpp::OneChangeTransitionModel::dPij_dt(), bpp::AbstractCodonBGCSubstitutionModel::getCodonsMulRate(), bpp::AbstractCodonDistanceSubstitutionModel::getCodonsMulRate(), bpp::BinarySubstitutionModel::getd2Pij_dt2(), bpp::F84::getd2Pij_dt2(), bpp::HKY85::getd2Pij_dt2(), bpp::TwoParameterBinarySubstitutionModel::getd2Pij_dt2(), bpp::EquiprobableSubstitutionModel::getd2Pij_dt2(), bpp::F81::getd2Pij_dt2(), bpp::JCnuc::getd2Pij_dt2(), bpp::K80::getd2Pij_dt2(), bpp::T92::getd2Pij_dt2(), bpp::TN93::getd2Pij_dt2(), bpp::JCprot::getd2Pij_dt2(), bpp::RE08::getd2Pij_dt2(), bpp::OneChangeTransitionModel::getd2Pij_dt2(), bpp::BinarySubstitutionModel::getdPij_dt(), bpp::F84::getdPij_dt(), bpp::HKY85::getdPij_dt(), bpp::TwoParameterBinarySubstitutionModel::getdPij_dt(), bpp::EquiprobableSubstitutionModel::getdPij_dt(), bpp::F81::getdPij_dt(), bpp::JCnuc::getdPij_dt(), bpp::K80::getdPij_dt(), bpp::T92::getdPij_dt(), bpp::TN93::getdPij_dt(), bpp::JCprot::getdPij_dt(), bpp::RE08::getdPij_dt(), bpp::OneChangeTransitionModel::getdPij_dt(), bpp::BinarySubstitutionModel::getPij_t(), bpp::F84::getPij_t(), bpp::HKY85::getPij_t(), bpp::TwoParameterBinarySubstitutionModel::getPij_t(), bpp::EquiprobableSubstitutionModel::getPij_t(), bpp::F81::getPij_t(), bpp::JCnuc::getPij_t(), bpp::K80::getPij_t(), bpp::T92::getPij_t(), bpp::TN93::getPij_t(), bpp::JCprot::getPij_t(), bpp::RE08::getPij_t(), bpp::OneChangeTransitionModel::getPij_t(), bpp::LegacyOptimizationTools::ScaleFunction::getValue(), bpp::LegacyOptimizationTools::optimizeTreeScale(), bpp::BinarySubstitutionModel::Pij_t(), bpp::F84::Pij_t(), bpp::HKY85::Pij_t(), bpp::TwoParameterBinarySubstitutionModel::Pij_t(), bpp::EquiprobableSubstitutionModel::Pij_t(), bpp::F81::Pij_t(), bpp::JCnuc::Pij_t(), bpp::K80::Pij_t(), bpp::T92::Pij_t(), bpp::TN93::Pij_t(), bpp::JCprot::Pij_t(), bpp::RE08::Pij_t(), bpp::OneChangeTransitionModel::Pij_t(), pow(), powi(), bpp::PhylogeneticsApplicationTools::printAnalysisInformation(), and bpp::gBGC::updateMatrices_().
◆ exp() [2/2]
|
inline |
Definition at line 909 of file ExtendedFloatEigen.h.
References bpp::ExtendedFloatEigen< R, C, EigenType >::exp().
◆ failureComputeWasCalled()
| void bpp::failureComputeWasCalled | ( | const std::type_info & | nodeType | ) |
Definition at line 51 of file DataFlow.cpp.
References prettyTypeName().
Referenced by bpp::NumericConstant< T >::compute(), bpp::NumericMutable< T >::compute(), and bpp::Sequence_DF::compute().
◆ failureDeltaNotDerivable()
| void bpp::failureDeltaNotDerivable | ( | const std::type_info & | contextNodeType | ) |
Definition at line 11 of file DataFlowCWiseComputing.cpp.
References prettyTypeName().
Referenced by bpp::CombineDeltaShifted< T >::derive().
◆ failureDependencyNumberMismatch()
| void bpp::failureDependencyNumberMismatch | ( | const std::type_info & | contextNodeType, |
| std::size_t | expectedSize, | ||
| std::size_t | givenSize | ||
| ) |
Definition at line 61 of file DataFlow.cpp.
References prettyTypeName(), and to_string().
Referenced by checkDependencyVectorMinSize(), and checkDependencyVectorSize().
◆ failureDependencyTypeMismatch()
| void bpp::failureDependencyTypeMismatch | ( | const std::type_info & | contextNodeType, |
| std::size_t | depIndex, | ||
| const std::type_info & | expectedType, | ||
| const std::type_info & | givenNodeType | ||
| ) |
Definition at line 74 of file DataFlow.cpp.
References prettyTypeName(), and to_string().
Referenced by checkNthDependencyIs(), bpp::ProbabilitiesFromMixedModel::create(), and bpp::ProbabilityFromMixedModel::create().
◆ failureEmptyDependency()
| void bpp::failureEmptyDependency | ( | const std::type_info & | contextNodeType, |
| std::size_t | depIndex | ||
| ) |
Definition at line 68 of file DataFlow.cpp.
References prettyTypeName(), and to_string().
Referenced by checkDependenciesNotNull(), and checkNthDependencyNotNull().
◆ failureNodeConversion()
| void bpp::failureNodeConversion | ( | const std::type_info & | handleType, |
| const Node_DF & | node | ||
| ) |
Definition at line 56 of file DataFlow.cpp.
References prettyTypeName().
Referenced by convertRef().
◆ failureNumericalDerivationNotConfigured()
| void bpp::failureNumericalDerivationNotConfigured | ( | ) |
Definition at line 16 of file DataFlowCWiseComputing.cpp.
Referenced by generateNumericalDerivative().
◆ generateNumericalDerivative() [1/2]
| ValueRef< NodeT > bpp::generateNumericalDerivative | ( | Context & | c, |
| const NumericalDerivativeConfiguration & | config, | ||
| NodeRef | dep, | ||
| const Dimension< DepT > & | depDim, | ||
| Dimension< NodeT > & | nodeDim, | ||
| B | buildNodeWithDep | ||
| ) |
Helper used to generate data flow expressions computing a numerical derivative.
For an expression e = f(x0,...,xn), which may be composed of multiple nodes. dep is one of the expression dependencies: exists i, dep is node xi. buildNodeWithDep(new_xi) should create the f(x0,...,new_xi,...,xn) node. It must duplicate the expression by replacing dep by new_xi. e must be a Value<NodeT>, and xi a Value<DepT>.
This function generates a new node de_ddep. de_ddep = g(delta,x0,...xn) = df/ddep value at x0,...,xn. delta is the shift between points of computation of f (numerical derivation). The pattern of computation points and delta are given by config. After creation, the pattern and node for delta cannot change, but delta value can.
Definition at line 2724 of file DataFlowCWiseComputing.h.
References bpp::ShiftDelta< T >::create(), bpp::CombineDeltaShifted< T >::create(), bpp::NumericalDerivativeConfiguration::delta, failureNumericalDerivationNotConfigured(), FivePoints, ThreePoints, and bpp::NumericalDerivativeConfiguration::type.
◆ generateNumericalDerivative() [2/2]
| ValueRef< NodeT > bpp::generateNumericalDerivative | ( | Context & | c, |
| const NumericalDerivativeConfiguration & | config, | ||
| NodeRef | dep, | ||
| const Dimension< NodeT > & | nodeDim, | ||
| B | buildNodeWithDep | ||
| ) |
Definition at line 2767 of file DataFlowCWiseComputing.h.
References bpp::CombineDeltaShifted< T >::create(), bpp::ShiftParameter::create(), bpp::NumericalDerivativeConfiguration::delta, failureNumericalDerivationNotConfigured(), FivePoints, ThreePoints, and bpp::NumericalDerivativeConfiguration::type.
◆ hash() [1/2]
| std::size_t bpp::hash | ( | const MatrixDimension & | dim | ) |
Definition at line 153 of file DataFlowNumeric.cpp.
References bpp::MatrixDimension::cols, combineHash(), and bpp::MatrixDimension::rows.
Referenced by bpp::ConstantZero< T >::hashAdditionalArguments(), bpp::ConstantOne< T >::hashAdditionalArguments(), bpp::NumericConstant< T >::hashAdditionalArguments(), and bpp::Sequence_DF::hashAdditionalArguments().
◆ hash() [2/2]
|
inline |
Definition at line 235 of file DataFlowNumeric.h.
◆ isTransitivelyDependentOn()
Check if searchedDependency if a transitive dependency of node.
Definition at line 254 of file DataFlow.cpp.
Referenced by bpp::CombineDeltaShifted< T >::derive().
◆ log() [1/2]
|
inline |
Definition at line 443 of file ExtendedFloat.h.
References bpp::ExtendedFloat::log().
Referenced by bpp::CWiseLog< T >::compute(), bpp::LogSumExp< R, T0, T1 >::compute(), bpp::UniformizationSubstitutionCount::computeCounts_(), bpp::PhyloStatistics::computeForSubtree_(), bpp::BranchLikelihood::computeLogLikelihood(), bpp::AbstractMutationProcess::detailedEvolve(), bpp::AbstractCodonAAFitnessSubstitutionModel::getCodonsMulRate(), bpp::AbstractCodonFitnessSubstitutionModel::getCodonsMulRate(), bpp::DRHomogeneousMixedTreeLikelihood::getLogLikelihood(), bpp::DRNonHomogeneousTreeLikelihood::getLogLikelihood(), bpp::DRHomogeneousTreeLikelihood::getLogLikelihood(), bpp::AlignedLikelihoodCalculation::getLogLikelihoodForASite(), bpp::DRHomogeneousMixedTreeLikelihood::getLogLikelihoodForASite(), bpp::DRNonHomogeneousTreeLikelihood::getLogLikelihoodForASite(), bpp::RHomogeneousTreeLikelihood::getLogLikelihoodForASite(), bpp::RNonHomogeneousTreeLikelihood::getLogLikelihoodForASite(), bpp::DRHomogeneousTreeLikelihood::getLogLikelihoodForASite(), bpp::DRHomogeneousMixedTreeLikelihood::getLogLikelihoodForASiteForARateClass(), bpp::DRNonHomogeneousTreeLikelihood::getLogLikelihoodForASiteForARateClass(), bpp::RHomogeneousMixedTreeLikelihood::getLogLikelihoodForASiteForARateClass(), bpp::RHomogeneousTreeLikelihood::getLogLikelihoodForASiteForARateClass(), bpp::RNonHomogeneousTreeLikelihood::getLogLikelihoodForASiteForARateClass(), bpp::DRHomogeneousTreeLikelihood::getLogLikelihoodForASiteForARateClass(), bpp::DRHomogeneousMixedTreeLikelihood::getLogLikelihoodForASiteForARateClassForAState(), bpp::DRNonHomogeneousTreeLikelihood::getLogLikelihoodForASiteForARateClassForAState(), bpp::RHomogeneousMixedTreeLikelihood::getLogLikelihoodForASiteForARateClassForAState(), bpp::RHomogeneousTreeLikelihood::getLogLikelihoodForASiteForARateClassForAState(), bpp::RNonHomogeneousTreeLikelihood::getLogLikelihoodForASiteForARateClassForAState(), bpp::DRHomogeneousTreeLikelihood::getLogLikelihoodForASiteForARateClassForAState(), bpp::AbstractDiscreteRatesAcrossSitesTreeLikelihood::getLogLikelihoodForASiteForAState(), bpp::ExtendedFloat::log(), and bpp::IOPhymlPairedSiteLikelihoods::readPairedSiteLikelihoods().
◆ log() [2/2]
|
inline |
Definition at line 902 of file ExtendedFloatEigen.h.
References bpp::ExtendedFloatEigen< R, C, EigenType >::log().
◆ makeConfiguredModelCollection()
|
inline |
Make a Collection of ConfiguredModel, from the models described in the SubstitutionProcess, and sharing ConfiguredParameters from a ParameterList.
Parameter names from the ParameterList may have "_suff" terminations, with suff matching the numbers of the models in the process.
Definition at line 262 of file ProcessTree.h.
References bpp::ParametrizableCollection< class >::addObject(), bpp::SubstitutionProcessInterface::getModel(), bpp::SubstitutionProcessInterface::getModelNumbers(), and bpp::TextTools::toString().
Referenced by bpp::ProcessTree::makeProcessTree().
◆ operator!=() [1/2]
|
inline |
Definition at line 298 of file DataFlowNumeric.h.
◆ operator!=() [2/2]
|
inline |
Definition at line 237 of file DataFlowNumeric.h.
◆ operator&()
| bool bpp::operator& | ( | DotOptions | a, |
| DotOptions | b | ||
| ) |
Definition at line 442 of file DataFlow.cpp.
◆ operator*() [1/4]
|
inline |
Definition at line 448 of file ExtendedFloat.h.
◆ operator*() [2/4]
|
inline |
Definition at line 961 of file ExtendedFloatEigen.h.
◆ operator*() [3/4]
|
inline |
Definition at line 1049 of file ExtendedFloatEigen.h.
◆ operator*() [4/4]
|
inline |
Definition at line 953 of file ExtendedFloatEigen.h.
References bpp::ExtendedFloatEigen< R, C, EigenType >::denorm_mul().
◆ operator+() [1/2]
|
inline |
Definition at line 458 of file ExtendedFloat.h.
◆ operator+() [2/2]
|
inline |
Definition at line 935 of file ExtendedFloatEigen.h.
◆ operator-() [1/2]
|
inline |
Definition at line 453 of file ExtendedFloat.h.
◆ operator-() [2/2]
|
inline |
Definition at line 944 of file ExtendedFloatEigen.h.
◆ operator/()
|
inline |
Definition at line 463 of file ExtendedFloat.h.
◆ operator<<()
|
inline |
Definition at line 431 of file ExtendedFloat.h.
References bpp::ExtendedFloat::exponent_part(), and bpp::ExtendedFloat::float_part().
◆ operator==() [1/2]
|
inline |
Definition at line 294 of file DataFlowNumeric.h.
References bpp::MatrixDimension::cols, and bpp::MatrixDimension::rows.
◆ operator==() [2/2]
|
inline |
Definition at line 236 of file DataFlowNumeric.h.
◆ operator|()
| DotOptions bpp::operator| | ( | DotOptions | a, |
| DotOptions | b | ||
| ) |
Definition at line 437 of file DataFlow.cpp.
◆ pow() [1/3]
|
inline |
Definition at line 469 of file ExtendedFloat.h.
References exp(), and bpp::ExtendedFloat::pow().
Referenced by bpp::CodonFromIndependentFrequencySet::CodonFromIndependentFrequencySet(), bpp::CodonFromUniqueFrequencySet::CodonFromUniqueFrequencySet(), bpp::CombineDeltaShifted< T >::compute(), bpp::CWiseConstantPow< T >::compute(), bpp::TreeTools::computeBootstrapValues(), bpp::PhyloTreeTools::computeBranchLengthsGrafen(), bpp::TreeTools::computeBranchLengthsGrafen(), bpp::LaplaceSubstitutionCount::computeCounts(), bpp::HierarchicalClustering::computeDistancesFromPair(), bpp::EquiprobableSubstitutionModel::d2Pij_dt2(), bpp::ExtendedFloat::denorm_pow(), bpp::ExtendedFloat::exp(), bpp::AbstractCodonAAFitnessSubstitutionModel::getCodonsMulRate(), bpp::RHomogeneousTreeLikelihood::getD2LogLikelihoodForASite(), bpp::RNonHomogeneousTreeLikelihood::getD2LogLikelihoodForASite(), bpp::EquiprobableSubstitutionModel::getd2Pij_dt2(), bpp::DRHomogeneousMixedTreeLikelihood::getLikelihood(), bpp::DRNonHomogeneousTreeLikelihood::getLikelihood(), bpp::DRHomogeneousTreeLikelihood::getLikelihood(), bpp::DRHomogeneousMixedTreeLikelihood::getSecondOrderDerivative(), bpp::DRNonHomogeneousTreeLikelihood::getSecondOrderDerivative(), bpp::DRHomogeneousTreeLikelihood::getSecondOrderDerivative(), bpp::CodonFromIndependentFrequencySet::updateFrequencies(), bpp::CodonFromUniqueFrequencySet::updateFrequencies(), bpp::RELAX::updateMatrices_(), and bpp::POMO::updateMatrices_().
◆ pow() [2/3]
|
inline |
Definition at line 915 of file ExtendedFloatEigen.h.
References bpp::ExtendedFloatEigen< R, C, EigenType >::denorm_pow(), and exp().
◆ pow() [3/3]
|
inline |
Definition at line 923 of file ExtendedFloatEigen.h.
References bpp::ExtendedFloatEigen< R, C, EigenType >::denorm_pow(), and exp().
Referenced by bpp::ExtendedFloatEigen< R, C, EigenType >::denorm_pow(), and bpp::ExtendedFloatEigen< R, C, EigenType >::exp().
◆ powi()
|
inline |
Definition at line 24 of file ExtendedFloat.h.
References exp().
Referenced by bpp::ExtendedFloat::lround().
◆ prettyTypeName()
| std::string bpp::prettyTypeName | ( | const std::type_info & | ti | ) |
Debug: return a readable name for a C++ type descriptor (from typeid operator).
Definition at line 43 of file DataFlow.cpp.
References demangle().
Referenced by checkNthDependencyIs(), checkRecreateWithoutDependencies(), bpp::Node_DF::description(), failureComputeWasCalled(), failureDeltaNotDerivable(), failureDependencyNumberMismatch(), failureDependencyTypeMismatch(), failureEmptyDependency(), and failureNodeConversion().
◆ recreateWithSubstitution()
| NodeRef bpp::recreateWithSubstitution | ( | Context & | c, |
| const NodeRef & | node, | ||
| const std::unordered_map< const Node_DF *, NodeRef > & | substitutions | ||
| ) |
Recreate node by transitively replacing dependencies according to substitutions mapping.
Definition at line 272 of file DataFlow.cpp.
References recreateWithSubstitution().
Referenced by recreateWithSubstitution().
◆ removeDependenciesIf()
| void bpp::removeDependenciesIf | ( | NodeRefVec & | deps, |
| Predicate | p | ||
| ) |
Definition at line 908 of file DataFlowNumeric.h.
Referenced by bpp::CWiseAdd< R, ReductionOf< T > >::create(), bpp::CWiseMean< R, ReductionOf< T >, ReductionOf< P > >::create(), and bpp::CWiseMul< R, ReductionOf< T > >::create().
◆ to_string() [1/3]
|
inline |
Definition at line 437 of file ExtendedFloat.h.
References bpp::ExtendedFloat::exponent_part(), bpp::ExtendedFloat::float_part(), and to_string().
Referenced by to_string().
◆ to_string() [2/3]
| std::string bpp::to_string | ( | const MatrixDimension & | dim | ) |
Definition at line 148 of file DataFlowNumeric.cpp.
References bpp::MatrixDimension::cols, bpp::MatrixDimension::rows, and to_string().
Referenced by bpp::numeric::checkDimensionIsSquare(), and to_string().
◆ to_string() [3/3]
| std::string bpp::to_string | ( | const NoDimension & | ) |
Definition at line 143 of file DataFlowNumeric.cpp.
Referenced by checkNthDependencyIs(), bpp::CWiseMean< R, ReductionOf< T >, ReductionOf< P > >::create(), bpp::numeric::debug(), bpp::ConfiguredDistribution::debugInfo(), bpp::ProbabilitiesFromDiscreteDistribution::debugInfo(), bpp::ConfiguredFrequencySet::debugInfo(), bpp::FrequenciesFromFrequencySet::debugInfo(), bpp::ConfiguredModel::debugInfo(), bpp::EquilibriumFrequenciesFromModel::debugInfo(), bpp::TransitionMatrixFromModel::debugInfo(), bpp::TransitionFunctionFromModel::debugInfo(), bpp::ProbabilitiesFromMixedModel::debugInfo(), bpp::ConfiguredSimplex::debugInfo(), bpp::FrequenciesFromSimplex::debugInfo(), bpp::ConfiguredTransitionMatrix::debugInfo(), bpp::EquilibriumFrequenciesFromTransitionMatrix::debugInfo(), bpp::TransitionMatrixFromTransitionMatrix::debugInfo(), bpp::CWiseFill< R, T >::debugInfo(), bpp::CWisePattern< R >::debugInfo(), bpp::CWiseMatching< R, ReductionOf< T > >::debugInfo(), bpp::CWiseCompound< R, ReductionOf< T > >::debugInfo(), bpp::CWiseApply< R, T, F >::debugInfo(), bpp::CWiseAdd< R, std::tuple< T0, T1 > >::debugInfo(), bpp::CWiseSub< R, std::tuple< T0, T1 > >::debugInfo(), bpp::CWiseAdd< R, ReductionOf< T > >::debugInfo(), bpp::CWiseAdd< R, T >::debugInfo(), bpp::CWiseMean< R, ReductionOf< T >, ReductionOf< P > >::debugInfo(), bpp::CWiseMean< R, ReductionOf< T >, P >::debugInfo(), bpp::CWiseMul< R, std::tuple< T0, T1 > >::debugInfo(), bpp::CWiseMul< R, ReductionOf< T > >::debugInfo(), bpp::CWiseDiv< R, std::tuple< T0, T1 > >::debugInfo(), bpp::CWiseNegate< T >::debugInfo(), bpp::CWiseInverse< T >::debugInfo(), bpp::CWiseLog< T >::debugInfo(), bpp::CWiseExp< T >::debugInfo(), bpp::CWiseConstantPow< T >::debugInfo(), bpp::MatrixProduct< R, T0, T1 >::debugInfo(), bpp::ShiftDelta< T >::debugInfo(), bpp::CombineDeltaShifted< T >::debugInfo(), bpp::ConstantZero< T >::debugInfo(), bpp::ConstantOne< T >::debugInfo(), bpp::Convert< R, F >::debugInfo(), bpp::Identity< R >::debugInfo(), bpp::ShiftParameter::debugInfo(), bpp::CondLikelihood::debugInfo(), bpp::ForwardHmmLikelihood_DF::debugInfo(), bpp::ForwardHmmDLikelihood_DF::debugInfo(), bpp::ForwardHmmD2Likelihood_DF::debugInfo(), bpp::BackwardHmmLikelihood_DF::debugInfo(), bpp::CombineDeltaShifted< T >::description(), bpp::ConfiguredParameter::description(), dotIdentifier(), failureDependencyNumberMismatch(), failureDependencyTypeMismatch(), failureEmptyDependency(), bpp::AbstractSubstitutionModel::getPij_t(), bpp::ModelPath::PathNode::to_string(), and to_string().
◆ to_string_with_precision()
| std::string bpp::to_string_with_precision | ( | const T | a_value, |
| const int | n = 6 |
||
| ) |
Output with given precision.
Definition at line 66 of file DataFlow.h.
◆ transitionFunctionDimension()
|
inline |
Definition at line 20 of file ForwardLikelihoodTree.h.
Referenced by bpp::ForwardLikelihoodTree::makeForwardLikelihoodAtEdge().
◆ transitionMatrixDimension()
|
inline |
Definition at line 19 of file Model.h.
Referenced by bpp::ForwardLikelihoodTree::makeForwardLikelihoodAtEdge(), and bpp::LikelihoodCalculationOnABranch::makeLikelihoods().
◆ writeDotEdge()
|
static |
Definition at line 492 of file DataFlow.cpp.
References bpp::Node_DF::color(), bpp::Node_DF::dependency(), dotIdentifier(), ShowDependencyIndex, and to().
Referenced by writeGraphStructure().
◆ writeDotNode()
|
static |
Definition at line 475 of file DataFlow.cpp.
References bpp::Node_DF::color(), bpp::Node_DF::debugInfo(), bpp::Node_DF::description(), DetailedNodeInfo, dotIdentifier(), dotLabelEscape(), bpp::Node_DF::isValid(), and bpp::Node_DF::shape().
Referenced by writeGraphStructure().
◆ writeGraphStructure()
|
static |
Definition at line 517 of file DataFlow.cpp.
References FollowUpwardLinks, writeDotEdge(), and writeDotNode().
Referenced by writeGraphToDot().
◆ writeGraphToDot() [1/2]
| void bpp::writeGraphToDot | ( | const std::string & | filename, |
| const std::vector< const Node_DF * > & | nodes, | ||
| DotOptions | opt | ||
| ) |
Write dataflow graph starting at nodes to file at filename, overwriting it.
Definition at line 571 of file DataFlow.cpp.
References writeGraphToDot().
◆ writeGraphToDot() [2/2]
| void bpp::writeGraphToDot | ( | std::ostream & | os, |
| const std::vector< const Node_DF * > & | nodes, | ||
| DotOptions | opt | ||
| ) |
Write dataflow graph starting at nodes to output stream.
Definition at line 564 of file DataFlow.cpp.
References writeGraphStructure().
Referenced by bpp::LikelihoodCalculationSingleProcess::makeLikelihoodsAtRoot_(), and writeGraphToDot().
Variable Documentation
◆ NodeX
| NumericConstant< char > bpp::NodeX | ( | 'X' | ) |
Definition at line 1475 of file DataFlowNumeric.h.
Referenced by bpp::CWiseApply< R, T, F >::derive(), and bpp::TransitionFunctionFromModel::derive().